SphereAdaSTEM model: spherical indexing and modeling¶
Yangkang Chen
Jan 24, 2024
This notebook is to show the main difference of SphereAdaSTEM compared to AdaSTEM. To be specific, only the gridding part is different, and all other functionalities are inherited from AdaSTEM class. For complete functionality, please refer to AdaSTEM Demo.
We will explore the same modeling task as AdaSTEM Demo did: Predict the abundance of Mallard (a bird species) based on environmental variables. The data were requested from eBird, a citizen science project for bird observation, and with some variable annotation.
import pandas as pd
import numpy as np
import random
from tqdm.auto import tqdm
import matplotlib.pyplot as plt
import matplotlib
import warnings
import pickle
import os
import h3pandas
pd.set_option('display.max_columns', None)
# warnings.filterwarnings('ignore')
%load_ext autoreload
%autoreload 2
Download data¶
Training/test data¶
Please download the sample data from:
Suppose now it's downloaded and saved as './Sample_data_Mallard.csv'
Alternatively, you can try other species like
- Alder Flycatcher: https://figshare.com/articles/dataset/Sample_data_Alder_Flycatcher_csv/24080751
- Short-eared Owl: https://figshare.com/articles/dataset/Sample_data_Short-eared_Owl_csv/24080742
- Eurasian Tree Sparrow: https://figshare.com/articles/dataset/Sample_data_Eurasian_Tree_Sparrow_csv/24080748
Caveat: These bird observation data are about 200MB each file.
data = pd.read_csv(f'./Sample_data_Mallard.csv')
data = data.drop('sampling_event_identifier', axis=1)
Prediction set¶
Prediction set are used to feed into a trained AdaSTEM model and make prediction: at some location, at some day of year, given the environmental variables, how many Mallard individual do I expected to observe?
The prediction set will be loaded after the model is trained.
Download the prediction set from: https://figshare.com/articles/dataset/Predset_2020_csv/24124980
Caveat: The file is about 700MB.
Get X and y¶
X = data.drop('count', axis=1)
y = data['count'].values
X.head()
| longitude | latitude | DOY | duration_minutes | Traveling | Stationary | Area | effort_distance_km | number_observers | obsvr_species_count | time_observation_started_minute_of_day | elevation_mean | slope_mean | eastness_mean | northness_mean | bio1 | bio2 | bio3 | bio4 | bio5 | bio6 | bio7 | bio8 | bio9 | bio10 | bio11 | bio12 | bio13 | bio14 | bio15 | bio16 | bio17 | bio18 | bio19 | closed_shrublands | cropland_or_natural_vegetation_mosaics | croplands | deciduous_broadleaf_forests | deciduous_needleleaf_forests | evergreen_broadleaf_forests | evergreen_needleleaf_forests | grasslands | mixed_forests | non_vegetated_lands | open_shrublands | permanent_wetlands | savannas | urban_and_built_up_lands | water_bodies | woody_savannas | entropy | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -83.472224 | 8.859308 | 22 | 300.0 | 1 | 0 | 0 | 4.828 | 5.0 | 34.0 | 476 | 7.555556 | 0.758156 | 0.036083 | -0.021484 | 24.883502 | 5.174890 | 59.628088 | 93.482247 | 30.529131 | 21.850519 | 8.678612 | 24.302626 | 26.536822 | 26.213334 | 23.864924 | 0.720487 | 0.127594 | 0.003156 | 0.001451 | 0.332425 | 0.026401 | 0.044218 | 0.260672 | 0.0 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.138889 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.0 | 0.777778 | 0.000000 | 0.000000 | 0.083333 | 0.000000 | 0.676720 |
| 1 | -2.687724 | 43.373323 | 290 | 90.0 | 1 | 0 | 0 | 0.570 | 2.0 | 151.0 | 1075 | 30.833336 | 3.376527 | 0.050544 | -0.099299 | 14.107917 | 5.224109 | 31.174167 | 376.543853 | 23.219421 | 6.461607 | 16.757814 | 9.048385 | 19.092725 | 19.236082 | 9.287841 | 0.171423 | 0.035598 | 0.004512 | 0.000081 | 0.084657 | 0.018400 | 0.030210 | 0.065007 | 0.0 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.333333 | 0.000000 | 0.000000 | 0.083333 | 0.0 | 0.0 | 0.000000 | 0.194444 | 0.027778 | 0.000000 | 0.361111 | 1.359063 |
| 2 | -89.884770 | 35.087255 | 141 | 10.0 | 0 | 1 | 0 | -1.000 | 2.0 | 678.0 | 575 | 91.777780 | 0.558100 | -0.187924 | -0.269078 | 17.396487 | 8.673912 | 28.688889 | 718.996078 | 32.948335 | 2.713938 | 30.234397 | 14.741099 | 13.759220 | 26.795849 | 7.747272 | 0.187089 | 0.031802 | 0.005878 | 0.000044 | 0.073328 | 0.026618 | 0.039616 | 0.059673 | 0.0 | 0.055556 | 0.0 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.305556 | 0.000000 | 0.0 | 0.0 | 0.000000 | 0.527778 | 0.000000 | 0.000000 | 0.111111 | 1.104278 |
| 3 | -99.216873 | 31.218510 | 104 | 9.0 | 1 | 0 | 0 | 0.805 | 2.0 | 976.0 | 657 | 553.166700 | 0.856235 | -0.347514 | -0.342971 | 20.740836 | 10.665164 | 35.409121 | 666.796919 | 35.909941 | 5.790119 | 30.119822 | 18.444353 | 30.734456 | 29.546417 | 11.701038 | 0.084375 | 0.025289 | 0.000791 | 0.000052 | 0.052866 | 0.004096 | 0.006064 | 0.015965 | 0.0 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.000000 | 0.000000 | 1.000000 | 0.000000 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | -0.000000 |
| 4 | -124.426730 | 43.065847 | 96 | 30.0 | 1 | 0 | 0 | 0.161 | 2.0 | 654.0 | 600 | 6.500000 | 0.491816 | -0.347794 | -0.007017 | 11.822340 | 6.766870 | 35.672897 | 396.157833 | 22.608788 | 3.639569 | 18.969219 | 8.184412 | 16.290802 | 17.258721 | 7.319234 | 0.144122 | 0.044062 | 0.000211 | 0.000147 | 0.089238 | 0.004435 | 0.004822 | 0.040621 | 0.0 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.361111 | 0.166667 | 0.000000 | 0.472222 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 1.020754 |
The features include:
spatial coordinates:
longitudeandlatitude(used for indexing, not actual training)
Temporal coordinate:
- day of year (
DOY): used for both indexing and training
- day of year (
Sampling parameters: These are parameters quantifying how the observation was made
duration_minutes: How long the observation was conducted- Observation protocol:
Traveling,Stationary, orArea effort_distance_km: how far have one travelednumber_observers: How many observers are there in the groupobsvr_species_count: How many bird species have the birder observed in the pasttime_observation_started_minute_of_day: When did the birder start birding
Topological features:
- Features of elevation:
elevation_mean - Features of slope magnitude and direction:
slope_mean,eastness_mean,northness_mean
- Features of elevation:
Bioclimate features:
- Summaries of yearly temperature and precipitation: from
bio1tobio19
- Summaries of yearly temperature and precipitation: from
Land cover features:
- Summaries of land cover, percentage of cover. For example,
closed_shrublands,urban_and_built_up_lands. entropy: Entropy of land cover
- Summaries of land cover, percentage of cover. For example,
As you can see, the environmental variables are almost static. However, dynamic features (e.g., daily temperature) is fully supported as input. See Tips for data types for details.
Now we can take a look at the target variable
plt.hist(np.log(y+1),bins=100)
plt.xlabel('log count')
plt.show()
zero_frac = np.sum(y==0)/len(y)
print(f'Percentage record with zero Mallard count: {zero_frac*100}%')
Percentage record with zero Mallard count: 83.09425%
The target data is extremely zero-inflated. 83% checklists have not Mallard observation. This poses the necessity of using hurdle model.
First thing first: Spatiotemporal train test split¶
from stemflow.model_selection import ST_train_test_split
X_train, X_test, y_train, y_test = ST_train_test_split(X, y,
Spatio1 = 'longitude',
Spatio2 = 'latitude',
Temporal1 = 'DOY',
Spatio_blocks_count = 50, Temporal_blocks_count=50,
random_state=42, test_size=0.3)
This makes sure that we have a held-out test set, so that the estimation will be reliable.
For details of parameter setting, see AdaSTEM Demo.
Train AdaSTEM hurdle model¶
from stemflow.model.SphereAdaSTEM import SphereAdaSTEM, SphereAdaSTEMClassifier, SphereAdaSTEMRegressor
from xgboost import XGBClassifier, XGBRegressor # remember to install xgboost if you use it as base model
from stemflow.model.Hurdle import Hurdle_for_AdaSTEM, Hurdle
We first import the models. Although some classes are not used, I imported them for complete showcase of function.
Here, we import the SphereAdaSTEM, SphereAdaSTEMClassifier and SphereAdaSTEMRegressor from stemflow.model.SphereAdaSTEM module.
## "hurdle in Ada"
model = SphereAdaSTEMRegressor(
base_model=Hurdle(
classifier=XGBClassifier(tree_method='hist',random_state=42, verbosity = 0, n_jobs=1),
regressor=XGBRegressor(tree_method='hist',random_state=42, verbosity = 0, n_jobs=1)
), # hurdel model for zero-inflated problem (e.g., count)
task='hurdle',
save_gridding_plot = True,
ensemble_fold=10, # data are modeled 10 times, each time with jitter and rotation in Quadtree algo
min_ensemble_required=7, # Only points covered by > 7 stixels will be predicted
grid_len_upper_threshold=2500, # force splitting if the grid length exceeds 2500 (km)
grid_len_lower_threshold=500, # stop splitting if the grid length fall short 500 (km)
temporal_start=1, # The next 4 params define the temporal sliding window
temporal_end=366,
temporal_step=25, # The window takes steps of 20 DOY (see AdaSTEM demo for details)
temporal_bin_interval=50, # Each window will contain data of 50 DOY
points_lower_threshold=50, # Only stixels with more than 50 samples are trained
Temporal1='DOY',
use_temporal_to_train=True, # In each stixel, whether 'DOY' should be a predictor
n_jobs=1
)
The parameter setting is mostly similar to AdaSTEM. The differences is that:
- We use
SphereAdaSTEMRegressoras regressor, and - We set
grid_len_upper_thresholdandgrid_len_lower_thresholdas real distance in kilometers, although the spatial columns arelongitudeandlatitude. - You must have a column named "longitude" and a column named "latitude" in you dataframe. The grid will be generated on the fly, and gridding will be based on real distance.
For other details of parameter setting, see AdaSTEM Demo.
We then fit the model by simply call:
# columns of X_train should only contain predictors and Spatio-temporal indicators ('longitude', 'latitude', Temporal1)
model.fit(X_train.reset_index(drop=True), y_train, verbosity=1)
Generating Ensemble: 100%|██████████| 10/10 [07:21<00:00, 44.14s/it]
training: 0%| | 0/10 [00:00<?, ?it/s]
SphereAdaSTEMRegressor(base_model=Hurdle(classifier=XGBClassifier(base_score=None,
booster=None,
callbacks=None,
colsample_bylevel=None,
colsample_bynode=None,
colsample_bytree=None,
early_stopping_rounds=None,
enable_categorical=False,
eval_metric=None,
feature_types=None,
gamma=None,
gpu_id=None,
grow_policy=None,
importance_type=None,
interaction_constraints=...
max_bin=None,
max_cat_threshold=None,
max_cat_to_onehot=None,
max_delta_step=None,
max_depth=None,
max_leaves=None,
min_child_weight=None,
missing=nan,
monotone_constraints=None,
n_estimators=100,
n_jobs=1,
num_parallel_tree=None,
predictor=None,
random_state=42, ...)),
grid_len_upper_threshold=2500, save_gridding_plot=True,
stixel_training_size_threshold=50, temporal_step=25)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
SphereAdaSTEMRegressor(base_model=Hurdle(classifier=XGBClassifier(base_score=None,
booster=None,
callbacks=None,
colsample_bylevel=None,
colsample_bynode=None,
colsample_bytree=None,
early_stopping_rounds=None,
enable_categorical=False,
eval_metric=None,
feature_types=None,
gamma=None,
gpu_id=None,
grow_policy=None,
importance_type=None,
interaction_constraints=...
max_bin=None,
max_cat_threshold=None,
max_cat_to_onehot=None,
max_delta_step=None,
max_depth=None,
max_leaves=None,
min_child_weight=None,
missing=nan,
monotone_constraints=None,
n_estimators=100,
n_jobs=1,
num_parallel_tree=None,
predictor=None,
random_state=42, ...)),
grid_len_upper_threshold=2500, save_gridding_plot=True,
stixel_training_size_threshold=50, temporal_step=25)Hurdle(classifier=XGBClassifier(base_score=None, booster=None, callbacks=None,
colsample_bylevel=None, colsample_bynode=None,
colsample_bytree=None,
early_stopping_rounds=None,
enable_categorical=False, eval_metric=None,
feature_types=None, gamma=None, gpu_id=None,
grow_policy=None, importance_type=None,
interaction_constraints=None,
learning_rate=None, max_bin=No...
feature_types=None, gamma=None, gpu_id=None,
grow_policy=None, importance_type=None,
interaction_constraints=None, learning_rate=None,
max_bin=None, max_cat_threshold=None,
max_cat_to_onehot=None, max_delta_step=None,
max_depth=None, max_leaves=None,
min_child_weight=None, missing=nan,
monotone_constraints=None, n_estimators=100,
n_jobs=1, num_parallel_tree=None, predictor=None,
random_state=42, ...))XGBClassifier(base_score=None, booster=None, callbacks=None,
colsample_bylevel=None, colsample_bynode=None,
colsample_bytree=None, early_stopping_rounds=None,
enable_categorical=False, eval_metric=None, feature_types=None,
gamma=None, gpu_id=None, grow_policy=None, importance_type=None,
interaction_constraints=None, learning_rate=None, max_bin=None,
max_cat_threshold=None, max_cat_to_onehot=None,
max_delta_step=None, max_depth=None, max_leaves=None,
min_child_weight=None, missing=nan, monotone_constraints=None,
n_estimators=100, n_jobs=1, num_parallel_tree=None,
predictor=None, random_state=42, ...)XGBClassifier(base_score=None, booster=None, callbacks=None,
colsample_bylevel=None, colsample_bynode=None,
colsample_bytree=None, early_stopping_rounds=None,
enable_categorical=False, eval_metric=None, feature_types=None,
gamma=None, gpu_id=None, grow_policy=None, importance_type=None,
interaction_constraints=None, learning_rate=None, max_bin=None,
max_cat_threshold=None, max_cat_to_onehot=None,
max_delta_step=None, max_depth=None, max_leaves=None,
min_child_weight=None, missing=nan, monotone_constraints=None,
n_estimators=100, n_jobs=1, num_parallel_tree=None,
predictor=None, random_state=42, ...)XGBRegressor(base_score=None, booster=None, callbacks=None,
colsample_bylevel=None, colsample_bynode=None,
colsample_bytree=None, early_stopping_rounds=None,
enable_categorical=False, eval_metric=None, feature_types=None,
gamma=None, gpu_id=None, grow_policy=None, importance_type=None,
interaction_constraints=None, learning_rate=None, max_bin=None,
max_cat_threshold=None, max_cat_to_onehot=None,
max_delta_step=None, max_depth=None, max_leaves=None,
min_child_weight=None, missing=nan, monotone_constraints=None,
n_estimators=100, n_jobs=1, num_parallel_tree=None, predictor=None,
random_state=42, ...)XGBRegressor(base_score=None, booster=None, callbacks=None,
colsample_bylevel=None, colsample_bynode=None,
colsample_bytree=None, early_stopping_rounds=None,
enable_categorical=False, eval_metric=None, feature_types=None,
gamma=None, gpu_id=None, grow_policy=None, importance_type=None,
interaction_constraints=None, learning_rate=None, max_bin=None,
max_cat_threshold=None, max_cat_to_onehot=None,
max_delta_step=None, max_depth=None, max_leaves=None,
min_child_weight=None, missing=nan, monotone_constraints=None,
n_estimators=100, n_jobs=1, num_parallel_tree=None, predictor=None,
random_state=42, ...)Plot QuadTree ensembles¶
model.gridding_plot
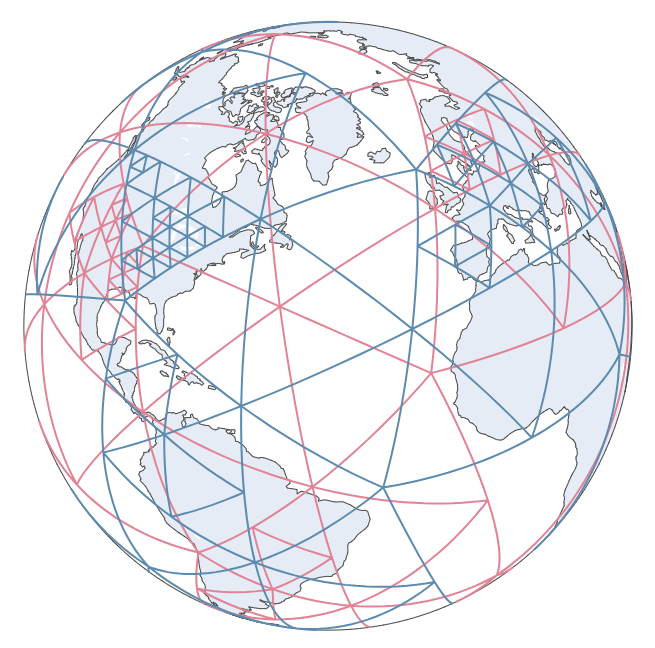
Here for an interactive plot Interactive spherical gridding plot.
with open('sphere_gridding_plot.html','w') as f:
f.write(model.gridding_plot.to_html())
This shows the 10 Quadtree ensembles we made. Region with higher data volume were split into smaller gird. The grid length is constrained between (500, 2500) kilometers.
Feature importances¶
After training the model, now we are interested in what features are important in bird abundance prediction
# Calcualte feature importance.
model.calculate_feature_importances()
# stixel-specific feature importance is saved in model.feature_importances_
top10_features = model.feature_importances_.iloc[:,1:].rank(axis=1).mean(axis=0).sort_values(ascending=False).head(10)
top10_features
slope_mean 34.278389 effort_distance_km 33.836596 elevation_mean 32.857154 northness_mean 30.856551 bio4 30.786596 eastness_mean 30.688630 duration_minutes 30.666340 obsvr_species_count 30.222590 bio8 29.530572 bio2 29.444051 dtype: float64
slope_mean, effort_distance_km, and elevation_mean are the top 3 predictors of Mallard abundance across the sampled space and time.
The feature importance ranking is similar to the results of AdaSTEM.
Evaluation¶
Now, we evaluate our overall model performance on the held-out test set:
pred = model.predict(X_test, verbosity=1)
predicting: 0%| | 0/10 [00:00<?, ?it/s]
The samples not predictable are output as np.nan:
perc = np.sum(np.isnan(pred.flatten()))/len(pred.flatten())
print(f'Percentage not predictable {round(perc*100, 2)}%')
Percentage not predictable 0.02%
More points are predictable compared to AdaSTEM model (0.02% unpredictable compared to ~1% unpredictable)! This is expected, as the Sphere indexing system enclose almost every corner of the earth.
We evaluate the performance using various metrics implemented in SphereAdaSTEM.eval_STEM_res or AdaSTEM.eval_STEM_res method:
pred_df = pd.DataFrame({
'y_true':y_test.flatten(),
'y_pred':np.where(pred.flatten()<0, 0, pred.flatten())
}).dropna()
SphereAdaSTEM.eval_STEM_res('hurdle', pred_df.y_true, pred_df.y_pred)
{'AUC': 0.7709769441127502,
'kappa': 0.40418935376349774,
'f1': 0.5315554903435247,
'precision': 0.4095122364777031,
'recall': 0.7572242253226733,
'average_precision': 0.35007503024002873,
'Spearman_r': 0.47773497480632715,
'Pearson_r': 0.20838678841242564,
'R2': -0.010718003793811492,
'MAE': 4.0526054794665,
'MSE': 1270.2122920088268,
'poisson_deviance_explained': 0.14793642063493806}
The model performance is similar compared to AdaSTEM model. The model performance should mainly be a result of parameters like ensemble_fold and points_lower_threshold, rather than the 2D or 3D gridding system.
Save model¶
We use pickle to save the model:
with open('./01.demo_adastem_model.pkl','wb') as f:
pickle.dump(model, f)
To load the model, do:
with open('./01.demo_adastem_model.pkl','rb') as f:
model = pickle.load(f)
Calculate memory usage¶
Finally, we are interested in how many disk space & memory our model consumed. stemflow is typically memory costly, especially when data volume and ensembles go up – the memory usage may also go up linearly. We do not currently provide solution for memory optimization. We well come PR to work on that.
# Calculate memory usage
model_size_G = round(os.path.getsize('./01.demo_adastem_model.pkl')/1024/1024/1024, 2)
training_data_memory_G = round(X.memory_usage().sum()/1024/1024/1024, 2)
# Calculate model info
true_hurdle_model_count = np.sum([1 if isinstance(model.model_dict[i].classifier, XGBClassifier) else 0 for i in model.model_dict])
dummy_model_count = len(model.model_dict) - true_hurdle_model_count
true_hurdle_model_perc = round(true_hurdle_model_count/len(model.model_dict) * 100, 2)
dummy_model_perc = round(dummy_model_count/len(model.model_dict) * 100, 2)
print(f"""
This AdaSTEM model have {len(model.model_dict)} trained based models in total.
Among them, {dummy_model_count} ({dummy_model_perc}%) are dummy models that always predict one class (because the input data labels are homogeneous).
Oppositely, {true_hurdle_model_count} ({true_hurdle_model_perc}%) are true hurdle models.
The input data consume {training_data_memory_G} G memory.
The model takes {model_size_G} G space on the disks.
""")
This AdaSTEM model have 6640 trained based models in total.
Among them, 1229 (18.51%) are dummy models that always predict one class (because the input data labels are homogeneous).
Oppositely, 5411 (81.49%) are true hurdle models.
The input data consume 0.15 G memory.
The model takes 1.7 G space on the disks.
Conclusion¶
Although we cannot directly compare the speed and performance of two modeling framework with different parameter settings, the results definitely show that, with similar gridding parameters:
For training:
AdaSTEMandSphereAdaSTEMhave similar speed.SphereAdaSTEMspends longer on gridding, but not significantly.
For prediction:
SphereAdaSTEMhas higher coverage, and therefore more samples in the test set can be predicted.- The prediction speed of
SphereAdaSTEMis almost double slower thanAdaSTEM. - The model performance is similar in terms of all metrics.
For memory use:
- The two model consume similar amount of memory.
My suggestion is that:
- For local modeling, you could use
AdaSTEMfor simplicity and understandability. - For global modeling, if you don't mind the difference in speed, you can use
SphereAdaSTEM. - For samples that distributed much towards the poles, definitely use
SphereAdaSTEM.
Please open an issue if you have any question
Cheers!
from watermark import watermark
print(watermark())
print(watermark(packages="stemflow,numpy,scipy,pandas,xgboost,tqdm,matplotlib,h3pandas,geopandas,scikit-learn"))
Last updated: 2024-01-24T21:47:37.851624+08:00 Python implementation: CPython Python version : 3.9.7 IPython version : 8.14.0 Compiler : Clang 11.1.0 OS : Darwin Release : 21.6.0 Machine : arm64 Processor : arm CPU cores : 8 Architecture: 64bit stemflow : 1.0.9.4 numpy : 1.24.3 scipy : 1.10.1 pandas : 2.0.3 xgboost : 1.7.6 tqdm : 4.65.0 matplotlib : 3.7.1 h3pandas : 0.2.4 geopandas : 0.11.1 scikit-learn: 0.0