A demo of AdaSTEM model¶
Yangkang Chen
Oct 10, 2025
This notebook is to provide a simple demonstration of how to use stemflow for AdaSTEM modeling.
For spherical indexing, see SphereAdaSTEM demo.
We will explore a modeling task: Predict the abundance of Mallard (a bird species) based on environmental variables. The data were requested from eBird, a citizen science project for bird observation, and with some variable annotation.
import os
import sys
import pandas as pd
import numpy as np
import random
from tqdm.auto import tqdm
import matplotlib.pyplot as plt
import matplotlib
import warnings
import pickle
pd.set_option('display.max_columns', None)
# warnings.filterwarnings('ignore')
%load_ext autoreload
%autoreload 2
The autoreload extension is already loaded. To reload it, use: %reload_ext autoreload
Download data¶
Training/test data¶
Please download the sample data from:
Suppose now it's downloaded and saved as './Sample_data_Mallard.csv'
Alternatively, you can try other species like
- Alder Flycatcher: https://figshare.com/articles/dataset/Sample_data_Alder_Flycatcher_csv/24080751
- Short-eared Owl: https://figshare.com/articles/dataset/Sample_data_Short-eared_Owl_csv/24080742
- Eurasian Tree Sparrow: https://figshare.com/articles/dataset/Sample_data_Eurasian_Tree_Sparrow_csv/24080748
Caveat: These bird observation data are about 200MB each file.
data = pd.read_csv(f'./Sample_data_Mallard.csv')
data = data.drop('sampling_event_identifier', axis=1)
Prediction set¶
Prediction set are used to feed into a trained AdaSTEM model and make prediction: at some location, at some day of year, given the environmental variables, how many Mallard individual do I expected to observe?
The prediction set will be loaded after the model is trained.
Download the prediction set from: https://figshare.com/articles/dataset/Predset_2020_csv/24124980
Caveat: The file is about 700MB.
Get X and y¶
X = data.drop('count', axis=1)
y = data[['count']]
X.head()
| longitude | latitude | DOY | duration_minutes | Traveling | Stationary | Area | effort_distance_km | number_observers | obsvr_species_count | time_observation_started_minute_of_day | elevation_mean | slope_mean | eastness_mean | northness_mean | bio1 | bio2 | bio3 | bio4 | bio5 | bio6 | bio7 | bio8 | bio9 | bio10 | bio11 | bio12 | bio13 | bio14 | bio15 | bio16 | bio17 | bio18 | bio19 | closed_shrublands | cropland_or_natural_vegetation_mosaics | croplands | deciduous_broadleaf_forests | deciduous_needleleaf_forests | evergreen_broadleaf_forests | evergreen_needleleaf_forests | grasslands | mixed_forests | non_vegetated_lands | open_shrublands | permanent_wetlands | savannas | urban_and_built_up_lands | water_bodies | woody_savannas | entropy | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -83.472224 | 8.859308 | 22 | 300.0 | 1 | 0 | 0 | 4.828 | 5.0 | 34.0 | 476 | 7.555556 | 0.758156 | 0.036083 | -0.021484 | 24.883502 | 5.174890 | 59.628088 | 93.482247 | 30.529131 | 21.850519 | 8.678612 | 24.302626 | 26.536822 | 26.213334 | 23.864924 | 0.720487 | 0.127594 | 0.003156 | 0.001451 | 0.332425 | 0.026401 | 0.044218 | 0.260672 | 0.0 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.138889 | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.0 | 0.777778 | 0.000000 | 0.000000 | 0.083333 | 0.000000 | 0.676720 |
| 1 | -2.687724 | 43.373323 | 290 | 90.0 | 1 | 0 | 0 | 0.570 | 2.0 | 151.0 | 1075 | 30.833336 | 3.376527 | 0.050544 | -0.099299 | 14.107917 | 5.224109 | 31.174167 | 376.543853 | 23.219421 | 6.461607 | 16.757814 | 9.048385 | 19.092725 | 19.236082 | 9.287841 | 0.171423 | 0.035598 | 0.004512 | 0.000081 | 0.084657 | 0.018400 | 0.030210 | 0.065007 | 0.0 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.333333 | 0.000000 | 0.000000 | 0.083333 | 0.0 | 0.0 | 0.000000 | 0.194444 | 0.027778 | 0.000000 | 0.361111 | 1.359063 |
| 2 | -89.884770 | 35.087255 | 141 | 10.0 | 0 | 1 | 0 | -1.000 | 2.0 | 678.0 | 575 | 91.777780 | 0.558100 | -0.187924 | -0.269078 | 17.396487 | 8.673912 | 28.688889 | 718.996078 | 32.948335 | 2.713938 | 30.234397 | 14.741099 | 13.759220 | 26.795849 | 7.747272 | 0.187089 | 0.031802 | 0.005878 | 0.000044 | 0.073328 | 0.026618 | 0.039616 | 0.059673 | 0.0 | 0.055556 | 0.0 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.305556 | 0.000000 | 0.0 | 0.0 | 0.000000 | 0.527778 | 0.000000 | 0.000000 | 0.111111 | 1.104278 |
| 3 | -99.216873 | 31.218510 | 104 | 9.0 | 1 | 0 | 0 | 0.805 | 2.0 | 976.0 | 657 | 553.166700 | 0.856235 | -0.347514 | -0.342971 | 20.740836 | 10.665164 | 35.409121 | 666.796919 | 35.909941 | 5.790119 | 30.119822 | 18.444353 | 30.734456 | 29.546417 | 11.701038 | 0.084375 | 0.025289 | 0.000791 | 0.000052 | 0.052866 | 0.004096 | 0.006064 | 0.015965 | 0.0 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.000000 | 0.000000 | 1.000000 | 0.000000 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | -0.000000 |
| 4 | -124.426730 | 43.065847 | 96 | 30.0 | 1 | 0 | 0 | 0.161 | 2.0 | 654.0 | 600 | 6.500000 | 0.491816 | -0.347794 | -0.007017 | 11.822340 | 6.766870 | 35.672897 | 396.157833 | 22.608788 | 3.639569 | 18.969219 | 8.184412 | 16.290802 | 17.258721 | 7.319234 | 0.144122 | 0.044062 | 0.000211 | 0.000147 | 0.089238 | 0.004435 | 0.004822 | 0.040621 | 0.0 | 0.000000 | 0.0 | 0.0 | 0.0 | 0.361111 | 0.166667 | 0.000000 | 0.472222 | 0.0 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 1.020754 |
The features include:
spatial coordinates:
longitudeandlatitude(used for indexing, not actual training)
Temporal coordinate:
- day of year (
DOY): used for both indexing and training
- day of year (
Sampling parameters: These are parameters quantifying how the observation was made
duration_minutes: How long the observation was conducted- Observation protocol:
Traveling,Stationary, orArea effort_distance_km: how far have one travelednumber_observers: How many observers are there in the groupobsvr_species_count: How many bird species have the birder observed in the pasttime_observation_started_minute_of_day: When did the birder start birding
Topological features:
- Features of elevation:
elevation_mean - Features of slope magnitude and direction:
slope_mean,eastness_mean,northness_mean
- Features of elevation:
Bioclimate features:
- Summaries of yearly temperature and precipitation: from
bio1tobio19
- Summaries of yearly temperature and precipitation: from
Land cover features:
- Summaries of land cover, percentage of cover. For example,
closed_shrublands,urban_and_built_up_lands. entropy: Entropy of land cover
- Summaries of land cover, percentage of cover. For example,
As you can see, the environmental variables are almost static. However, dynamic features (e.g., daily temperature) is fully supported as input. See Tips for data types for details.
Now we can take a look at the target variable
plt.hist(np.log(y+1),bins=100)
plt.xlabel('log count')
plt.show()
zero_frac = np.sum(y==0)/len(y)
print(f'Percentage record with zero Mallard count: {zero_frac*100}%')
Percentage record with zero Mallard count: count 83.09425 dtype: float64%
The target data is extremely zero-inflated. 83% checklists have not Mallard observation. This poses the necessity of using hurdle model.
First thing first: Spatiotemporal train test split¶
from stemflow.model_selection import ST_train_test_split
X_train, X_test, y_train, y_test = ST_train_test_split(X, y,
Spatio1 = 'longitude',
Spatio2 = 'latitude',
Temporal1 = 'DOY',
Spatio_blocks_count = 50, Temporal_blocks_count=50,
random_state=42, test_size=0.3)
Here we used a spatiotemporal train-test-spit to split the data into different blocks.
As shown, longitude and latitude are split into 50 bins separately, and DOY is split into 50 bins as well.
We then randomly select 30% of the spatiotemporal blocks as test data, and the rest as training data.
That is, if the data X have longitude ranging from (-180, 180), latitude ranging from (-90, 90), and whole year data (1, 366), each block will approximately contain data of 7.2 longitude (about 800km), 3.6 latitude (about 400km), and 7 days, which approximately catch the spatiotemporal scale of bird migration. These are rough estimates to get a sense of the scale. Note: the actual geometry (in meter) of 1-degree longitude will change by the latitudinal positions. In that case, you may consider projecting your data to and equal-area coordinates. In our case, we believe that the bias induced by coordinate system is relatively low, considering the size of our grids (and this is the coordinate system used in the original AdaSTEM paper, although it is a apparent issue).
The underlying interpretation is that: the generalization performance to adjacent (800km, 400km, 7days) block is [TEST RESULT]. As the bin count getting larger, the bin size get smaller, and your estimation becoming more radical and optimistic.
You can input any coordinate and set different bins as long as they match your model assumption.
Train AdaSTEM hurdle model¶
from stemflow.model.AdaSTEM import AdaSTEM, AdaSTEMClassifier, AdaSTEMRegressor
from xgboost import XGBClassifier, XGBRegressor # remember to install xgboost if you use it as base model
from stemflow.model.Hurdle import Hurdle_for_AdaSTEM, Hurdle
We first import the models. Although some classes are not used, I imported them for complete showcase of function.
## "hurdle in Ada"
model = AdaSTEMRegressor(
base_model=Hurdle(
classifier=XGBClassifier(tree_method='hist',random_state=42, verbosity = 0, n_jobs=1),
regressor=XGBRegressor(tree_method='hist',random_state=42, verbosity = 0, n_jobs=1)
), # hurdel model for zero-inflated problem (e.g., count)
task='hurdle',
save_gridding_plot = True,
ensemble_fold=10, # data are modeled 10 times, each time with jitter and rotation in Quadtree algo
min_ensemble_required=7, # Only points covered by > 7 ensembles will be predicted
grid_len_upper_threshold=25, # force splitting if the grid length exceeds 25
grid_len_lower_threshold=5, # stop splitting if the grid length fall short 5
temporal_start=1, # The next 4 params define the temporal sliding window
temporal_end=366,
temporal_step=25, # The window takes steps of 20 DOY (see AdaSTEM demo for details)
temporal_bin_interval=50, # Each window will contain data of 50 DOY
points_lower_threshold=50, # Only stixels with more than 50 samples are trained
Spatio1='longitude', # The next three params define the name of
Spatio2='latitude', # spatial coordinates shown in the dataframe
Temporal1='DOY',
use_temporal_to_train=True, # In each stixel, whether 'DOY' should be a predictor
n_jobs=1, # Not using parallel computing
random_state=42, # The random state makes the gridding process reproducible
lazy_loading=True # Using lazy loading for large ensemble amount (e.g., >20 ensembles).
# -- Each trained ensemble will be saved into disk and will only be loaded if needed (e.g. for prediction).
)
Here we used the Hurdle model as based model of AdaSTEMRegressor. For more discussion on modeling framework, see Tips for different tasks and Model structure of AdaSTEM and Hurdle.
During the "split-apply-combine" process, the data is first chunked into temporal windows by sliding window approach:
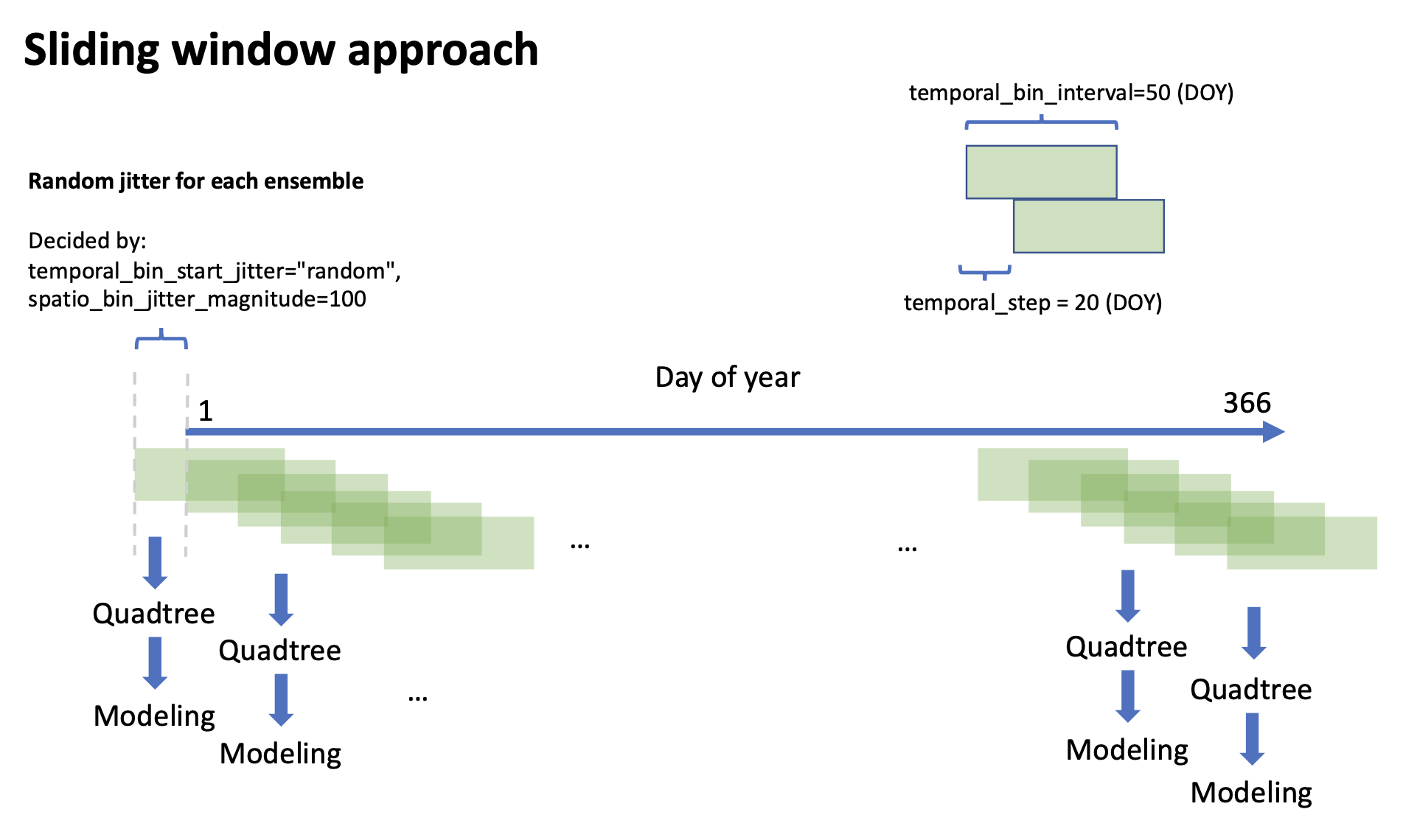
Then, for each temporal window, we split the data into spatial grids:
We choose adaptive grid size between 5 unit and 25 unit because these parameters was used in the original AdaSTEM paper. Large gird size will reduce the model performance (underfitting & over-extrapolation), and small gird size is likely overfitting to local condition. Likewise, we choose temporal window with size of 50 DOY and step of 25 DOY for that this timescale captures the onset and dynamics of migration. For more discussion on parameters please see Optimizing stixel size. We ask that only stixels with more than 50 samples are trained, to avoid incomplete sampling during bird survey. This is also recommended in the AdaSTEM paper. For rare/hard-to-observe species, the value should be set larger.
You could also use 3D spherical indexing, which is unbiased towards poles: SphereAdaSTEM demo.
We then fit the model by simply call:
# columns of X_train should only contain predictors and Spatio-temporal indicators (Spatio1, Spatio2, Temporal1)
model.fit(X_train, y_train, verbosity=1)
Generating Ensembles: 100%|██████████| 10/10 [00:17<00:00, 1.72s/it] Training: 100%|██████████| 10/10 [09:40<00:00, 58.03s/it]
AdaSTEMRegressor(base_model=Hurdle(classifier=XGBClassifier(base_score=None,
booster=None,
callbacks=None,
colsample_bylevel=None,
colsample_bynode=None,
colsample_bytree=None,
device=None,
early_stopping_rounds=None,
enable_categorical=False,
eval_metric=None,
feature_types=None,
feature_weights=None,
gamma=None,
grow_policy=None,
importance_type=None,
interacti...
n_estimators=None,
n_jobs=1,
num_parallel_tree=None, ...)),
lazy_loading=True,
lazy_loading_dir='/var/folders/j_/w98j87ps77j1k44x5fhz3dz80000gn/T/stemflow_model_62SctKDDhGPC18g1',
plot_xlims=(-179.8730564, 178.7306634),
plot_ylims=(-66.6730616, 73.0109413), random_state=42,
save_gridding_plot=True, stixel_training_size_threshold=50,
task='hurdle', temporal_step=25)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
AdaSTEMRegressor(base_model=Hurdle(classifier=XGBClassifier(base_score=None,
booster=None,
callbacks=None,
colsample_bylevel=None,
colsample_bynode=None,
colsample_bytree=None,
device=None,
early_stopping_rounds=None,
enable_categorical=False,
eval_metric=None,
feature_types=None,
feature_weights=None,
gamma=None,
grow_policy=None,
importance_type=None,
interacti...
n_estimators=None,
n_jobs=1,
num_parallel_tree=None, ...)),
lazy_loading=True,
lazy_loading_dir='/var/folders/j_/w98j87ps77j1k44x5fhz3dz80000gn/T/stemflow_model_62SctKDDhGPC18g1',
plot_xlims=(-179.8730564, 178.7306634),
plot_ylims=(-66.6730616, 73.0109413), random_state=42,
save_gridding_plot=True, stixel_training_size_threshold=50,
task='hurdle', temporal_step=25)Hurdle(classifier=XGBClassifier(base_score=None, booster=None, callbacks=None,
colsample_bylevel=None, colsample_bynode=None,
colsample_bytree=None, device=None,
early_stopping_rounds=None,
enable_categorical=False, eval_metric=None,
feature_types=None, feature_weights=None,
gamma=None, grow_policy=None,
importance_type=None,
interaction_constraints=None,
learning...
feature_types=None, feature_weights=None,
gamma=None, grow_policy=None,
importance_type=None,
interaction_constraints=None, learning_rate=None,
max_bin=None, max_cat_threshold=None,
max_cat_to_onehot=None, max_delta_step=None,
max_depth=None, max_leaves=None,
min_child_weight=None, missing=nan,
monotone_constraints=None, multi_strategy=None,
n_estimators=None, n_jobs=1,
num_parallel_tree=None, ...))XGBClassifier(base_score=None, booster=None, callbacks=None,
colsample_bylevel=None, colsample_bynode=None,
colsample_bytree=None, device=None, early_stopping_rounds=None,
enable_categorical=False, eval_metric=None, feature_types=None,
feature_weights=None, gamma=None, grow_policy=None,
importance_type=None, interaction_constraints=None,
learning_rate=None, max_bin=None, max_cat_threshold=None,
max_cat_to_onehot=None, max_delta_step=None, max_depth=None,
max_leaves=None, min_child_weight=None, missing=nan,
monotone_constraints=None, multi_strategy=None, n_estimators=None,
n_jobs=1, num_parallel_tree=None, ...)XGBClassifier(base_score=None, booster=None, callbacks=None,
colsample_bylevel=None, colsample_bynode=None,
colsample_bytree=None, device=None, early_stopping_rounds=None,
enable_categorical=False, eval_metric=None, feature_types=None,
feature_weights=None, gamma=None, grow_policy=None,
importance_type=None, interaction_constraints=None,
learning_rate=None, max_bin=None, max_cat_threshold=None,
max_cat_to_onehot=None, max_delta_step=None, max_depth=None,
max_leaves=None, min_child_weight=None, missing=nan,
monotone_constraints=None, multi_strategy=None, n_estimators=None,
n_jobs=1, num_parallel_tree=None, ...)XGBRegressor(base_score=None, booster=None, callbacks=None,
colsample_bylevel=None, colsample_bynode=None,
colsample_bytree=None, device=None, early_stopping_rounds=None,
enable_categorical=False, eval_metric=None, feature_types=None,
feature_weights=None, gamma=None, grow_policy=None,
importance_type=None, interaction_constraints=None,
learning_rate=None, max_bin=None, max_cat_threshold=None,
max_cat_to_onehot=None, max_delta_step=None, max_depth=None,
max_leaves=None, min_child_weight=None, missing=nan,
monotone_constraints=None, multi_strategy=None, n_estimators=None,
n_jobs=1, num_parallel_tree=None, ...)XGBRegressor(base_score=None, booster=None, callbacks=None,
colsample_bylevel=None, colsample_bynode=None,
colsample_bytree=None, device=None, early_stopping_rounds=None,
enable_categorical=False, eval_metric=None, feature_types=None,
feature_weights=None, gamma=None, grow_policy=None,
importance_type=None, interaction_constraints=None,
learning_rate=None, max_bin=None, max_cat_threshold=None,
max_cat_to_onehot=None, max_delta_step=None, max_depth=None,
max_leaves=None, min_child_weight=None, missing=nan,
monotone_constraints=None, multi_strategy=None, n_estimators=None,
n_jobs=1, num_parallel_tree=None, ...)Plot QuadTree ensembles¶
model.gridding_plot
This shows the 10 Quadtree ensembles we made. Region with higher data volume were split into smaller gird. The grid length is constrained between (5, 25) unit.
If you set n_jobs > 1, this grid plotting may not work. But you can plot it in another way. See below.
Take a look at the ensemble grids:
model.ensemble_df
| stixel_indexes | stixel_width | stixel_height | stixel_checklist_count | stixel_calibration_point(transformed) | rotation | ensemble_index | DOY_start | DOY_end | unique_stixel_id | stixel_calibration_point_transformed_left_bound | stixel_calibration_point_transformed_lower_bound | stixel_calibration_point_transformed_right_bound | stixel_calibration_point_transformed_upper_bound | calibration_point_x_jitter | calibration_point_y_jitter | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 5 | 22.239557 | 22.239557 | 83 | (80.095045, -167.156232) | 0.0 | 0 | -8.9 | 41.1 | 0_0_5 | 80.095045 | -167.156232 | 102.334602 | -144.916675 | 257.478369 | -167.201840 |
| 1 | 15 | 22.239557 | 22.239557 | 667 | (146.813715, -167.156232) | 0.0 | 0 | -8.9 | 41.1 | 0_0_15 | 146.813715 | -167.156232 | 169.053272 | -144.916675 | 257.478369 | -167.201840 |
| 2 | 19 | 22.239557 | 22.239557 | 83 | (102.334602, -122.677118) | 0.0 | 0 | -8.9 | 41.1 | 0_0_19 | 102.334602 | -122.677118 | 124.574159 | -100.437561 | 257.478369 | -167.201840 |
| 3 | 24 | 22.239557 | 22.239557 | 2193 | (124.574159, -144.916675) | 0.0 | 0 | -8.9 | 41.1 | 0_0_24 | 124.574159 | -144.916675 | 146.813716 | -122.677118 | 257.478369 | -167.201840 |
| 4 | 25 | 22.239557 | 22.239557 | 1362 | (124.574159, -122.677118) | 0.0 | 0 | -8.9 | 41.1 | 0_0_25 | 124.574159 | -122.677118 | 146.813716 | -100.437561 | 257.478369 | -167.201840 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 9463 | 124 | 11.273464 | 11.273464 | 73 | (354.138342, 293.79397) | 81.0 | 9 | 344.0 | 394.0 | 9_14_124 | 354.138342 | 293.793970 | 365.411806 | 305.067434 | 323.648072 | 193.096725 |
| 9464 | 125 | 11.273464 | 11.273464 | 250 | (342.864877, 305.067434) | 81.0 | 9 | 344.0 | 394.0 | 9_14_125 | 342.864877 | 305.067434 | 354.138341 | 316.340898 | 323.648072 | 193.096725 |
| 9465 | 126 | 11.273464 | 11.273464 | 513 | (342.864877, 316.340899) | 81.0 | 9 | 344.0 | 394.0 | 9_14_126 | 342.864877 | 316.340899 | 354.138341 | 327.614363 | 323.648072 | 193.096725 |
| 9466 | 127 | 11.273464 | 11.273464 | 152 | (354.138342, 305.067434) | 81.0 | 9 | 344.0 | 394.0 | 9_14_127 | 354.138342 | 305.067434 | 365.411806 | 316.340898 | 323.648072 | 193.096725 |
| 9467 | 128 | 11.273464 | 11.273464 | 83 | (354.138342, 316.340899) | 81.0 | 9 | 344.0 | 394.0 | 9_14_128 | 354.138342 | 316.340899 | 365.411806 | 327.614363 | 323.648072 | 193.096725 |
9468 rows × 16 columns
Feature importances¶
After training the model, now we are interested in what features are important in bird abundance prediction
# Calcualte feature importance.
model.calculate_feature_importances()
# stixel-specific feature importance is saved in model.feature_importances_
The feature importances for each stixel are calculated:
model.feature_importances_.sample(5)
| stixel_index | DOY | duration_minutes | Traveling | Stationary | Area | effort_distance_km | number_observers | obsvr_species_count | time_observation_started_minute_of_day | elevation_mean | slope_mean | eastness_mean | northness_mean | bio1 | bio2 | bio3 | bio4 | bio5 | bio6 | bio7 | bio8 | bio9 | bio10 | bio11 | bio12 | bio13 | bio14 | bio15 | bio16 | bio17 | bio18 | bio19 | closed_shrublands | cropland_or_natural_vegetation_mosaics | croplands | deciduous_broadleaf_forests | deciduous_needleleaf_forests | evergreen_broadleaf_forests | evergreen_needleleaf_forests | grasslands | mixed_forests | non_vegetated_lands | open_shrublands | permanent_wetlands | savannas | urban_and_built_up_lands | water_bodies | woody_savannas | entropy | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4956 | 5_6_96 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 2.040816e-02 | 0.020408 | 0.020408 | 0.020408 |
| 4336 | 4_9_126 | 0.028618 | 0.017718 | 0.056598 | 0.000000 | 0.000000 | 0.096100 | 0.009810 | 0.016755 | 0.012053 | 0.025475 | 0.048454 | 0.013461 | 0.027011 | 0.021664 | 0.041251 | 0.041686 | 0.016014 | 0.023750 | 0.017799 | 0.030231 | 0.012046 | 0.018057 | 0.018429 | 0.014066 | 0.016464 | 0.022564 | 0.015788 | 0.023004 | 0.020536 | 0.013779 | 0.021880 | 0.024241 | 0.000000 | 0.012348 | 0.013771 | 0.010456 | 0.000000 | 0.000000 | 0.008985 | 0.027858 | 0.040533 | 0.000000 | 0.000000 | 0.008911 | 0.051332 | 1.047411e-02 | 0.018010 | 0.017028 | 0.014992 |
| 6827 | 7_7_119 | 0.022044 | 0.025573 | 0.009145 | 0.000000 | 0.000000 | 0.012320 | 0.030115 | 0.036335 | 0.034802 | 0.063634 | 0.071332 | 0.019725 | 0.015117 | 0.022932 | 0.015261 | 0.007614 | 0.029310 | 0.008532 | 0.021710 | 0.026769 | 0.012171 | 0.004301 | 0.023688 | 0.026595 | 0.014087 | 0.040295 | 0.035353 | 0.012911 | 0.032106 | 0.011089 | 0.025771 | 0.006707 | 0.000000 | 0.000000 | 0.021620 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.019535 | 0.000000 | 0.000000 | 0.044684 | 0.000000 | 0.005050 | 1.800326e-01 | 0.001038 | 0.000000 | 0.010697 |
| 1718 | 1_11_92 | 0.014675 | 0.027601 | 0.008597 | 0.000000 | 0.000000 | 0.005647 | 0.000043 | 0.007494 | 0.019846 | 0.008228 | 0.066476 | 0.021662 | 0.007768 | 0.010531 | 0.035809 | 0.002934 | 0.007115 | 0.001518 | 0.014406 | 0.000000 | 0.014617 | 0.049055 | 0.005082 | 0.009511 | 0.005541 | 0.134597 | 0.002774 | 0.001702 | 0.014708 | 0.058732 | 0.075688 | 0.001992 | 0.000000 | 0.022687 | 0.000000 | 0.000000 | 0.000000 | 0.258600 | 0.006077 | 0.056022 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.004704 | 1.391074e-07 | 0.000000 | 0.010357 | 0.007203 |
| 7451 | 8_3_47 | 0.086313 | 0.009195 | 0.000143 | 0.000000 | 0.000000 | 0.016068 | 0.003115 | 0.024509 | 0.084288 | 0.005883 | 0.032775 | 0.004218 | 0.161167 | 0.007198 | 0.002697 | 0.002313 | 0.206739 | 0.000002 | 0.002423 | 0.151574 | 0.040709 | 0.005737 | 0.005891 | 0.001243 | 0.008539 | 0.004460 | 0.006967 | 0.000000 | 0.000000 | 0.000950 | 0.000937 | 0.035706 | 0.000000 | 0.002808 | 0.018816 | 0.001829 | 0.000000 | 0.007334 | 0.000000 | 0.011303 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.021598 | 6.805562e-04 | 0.000000 | 0.016787 | 0.007084 |
The stixel index naming follows {temporal_bin_index}_{ensemble_fold_index}_{grid_index}. For example, 17_7_103 means the 17th temporal bin, 7th ensemble, and 103th gird. The digits shown are feature importance calculated by the .feature_importances_ function of base model. Make sure your base model have one.
We can try to calculate the overall feature importance by average the ranking (or average the value directly). Notice: the assumption of this averaging is that the feature importance are homogeneous among different scales (that Quadtree generated), which may not be true.
top10_features = model.feature_importances_.iloc[:,1:].rank(axis=1).mean(axis=0).sort_values(ascending=False).head(10)
top10_features
slope_mean 33.554347 effort_distance_km 33.091942 elevation_mean 32.077292 duration_minutes 31.268340 eastness_mean 31.017397 northness_mean 30.639718 obsvr_species_count 30.347948 DOY 29.939459 bio4 29.460142 bio2 28.929926 dtype: float64
Looks like slope_mean, effort_distance_km, and elevation_mean are the top 3 predictors of Mallard abundance across the sampled space and time. They indicate that sampling parameters and topography may play important role here.
Noteworthy, it is not saying that other features, like temperate, are not important. We split the data into spatiotemporal blocks, and these feature importances can only represent what is important for predicting the abundance within the stixel (at the local level), not across them.
Now we want to visualize the feature importance by mapping them to spatiotemporal points. Our query points are constructed with 1-degree length and 7-days interval:
# make query points
Spatio_var1 = np.arange(-180, 180, 1)
Spatio_var2 = np.arange(-90, 90, 1)
Temporal_var1 = np.arange(1, 366, 7)
new_Spatio_var1, new_Spatio_var2, new_Temporal_var1 = np.meshgrid(Spatio_var1, Spatio_var2, Temporal_var1)
Sample_ST_df = pd.DataFrame(
{
model.Temporal1: new_Temporal_var1.flatten(),
model.Spatio1: new_Spatio_var1.flatten(),
model.Spatio2: new_Spatio_var2.flatten(),
}
)
# Assign the feature importance to spatio-temporal points of interest
importances_by_points = model.assign_feature_importances_by_points(Sample_ST_df, verbosity=1, n_jobs=1)
Querying ensembles: 100%|██████████| 10/10 [02:55<00:00, 17.59s/it]
importances_by_points.head()
| DOY | longitude | latitude | DOY_predictor | duration_minutes | Traveling | Stationary | Area | effort_distance_km | number_observers | obsvr_species_count | time_observation_started_minute_of_day | elevation_mean | slope_mean | eastness_mean | northness_mean | bio1 | bio2 | bio3 | bio4 | bio5 | bio6 | bio7 | bio8 | bio9 | bio10 | bio11 | bio12 | bio13 | bio14 | bio15 | bio16 | bio17 | bio18 | bio19 | closed_shrublands | cropland_or_natural_vegetation_mosaics | croplands | deciduous_broadleaf_forests | deciduous_needleleaf_forests | evergreen_broadleaf_forests | evergreen_needleleaf_forests | grasslands | mixed_forests | non_vegetated_lands | open_shrublands | permanent_wetlands | savannas | urban_and_built_up_lands | water_bodies | woody_savannas | entropy | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 482780 | 22 | -71 | -65 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 |
| 482886 | 22 | -69 | -65 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 |
| 482939 | 22 | -68 | -65 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 |
| 482992 | 22 | -67 | -65 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 |
| 501807 | 22 | -72 | -64 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 | 0.020408 |
# top 10 important variables
top_10_important_vars = importances_by_points[[
i for i in importances_by_points.columns if not i in ['DOY','longitude','latitude']
]].mean().sort_values(ascending=False).head(10)
print(top_10_important_vars)
DOY_predictor 0.044791 duration_minutes 0.037571 effort_distance_km 0.031669 bio4 0.029751 elevation_mean 0.029368 eastness_mean 0.027819 slope_mean 0.027661 northness_mean 0.027527 obsvr_species_count 0.025845 bio2 0.024091 dtype: float64
We see that feature importances assigned to spatiotemporal points have similar but different top-ranking features. This is expected, for different methods are used.
Ploting the feature importances by vairable names¶
We continue to visualize these feature important of dynamics:
from stemflow.utils.plot_gif import make_sample_gif
# make spatio-temporal GIF for top 3 variables
for var_ in top_10_important_vars.index[:3]:
make_sample_gif(importances_by_points, f'./FTR_IPT_{var_}.gif',
col=var_, log_scale = False,
Spatio1='longitude', Spatio2='latitude', Temporal1='DOY',
figsize=(18,9), xlims=(-180, 180), ylims=(-90,90), grid=True,
xtick_interval=20, ytick_interval=20,
lng_size = 360, lat_size = 180, dpi=100, fps=10)
0.0001 0.5281177375997815 Processing frame 53/53 Animation saved successfully! 0.0001 0.537394547036716 Processing frame 53/53 Animation saved successfully! 0.0001 0.42745354771614075 Processing frame 53/53 Animation saved successfully!
top_10_important_vars.index[:3]
Index(['DOY_predictor', 'duration_minutes', 'effort_distance_km'], dtype='object')
The feature importance for vairable DOY_predictor:
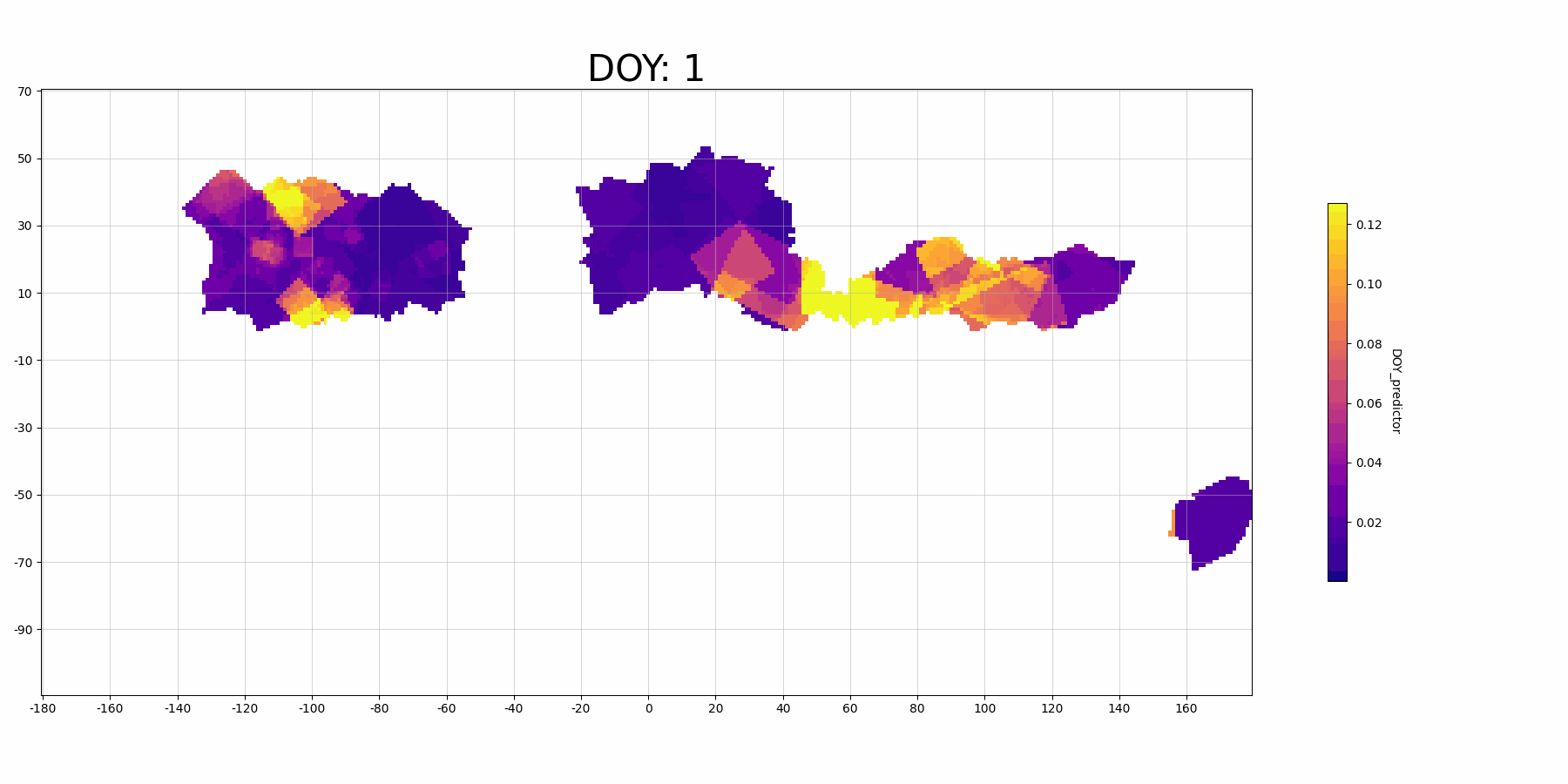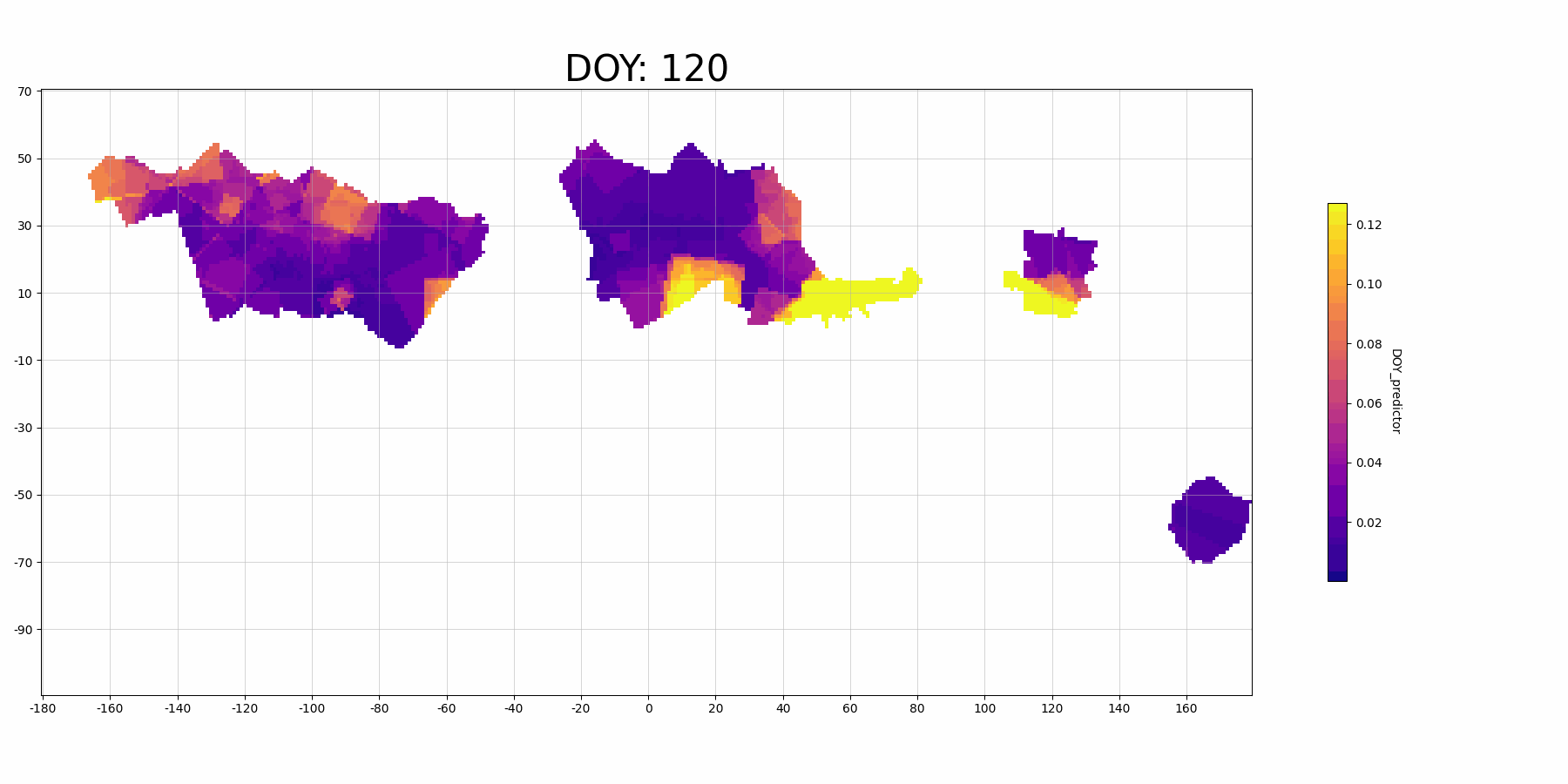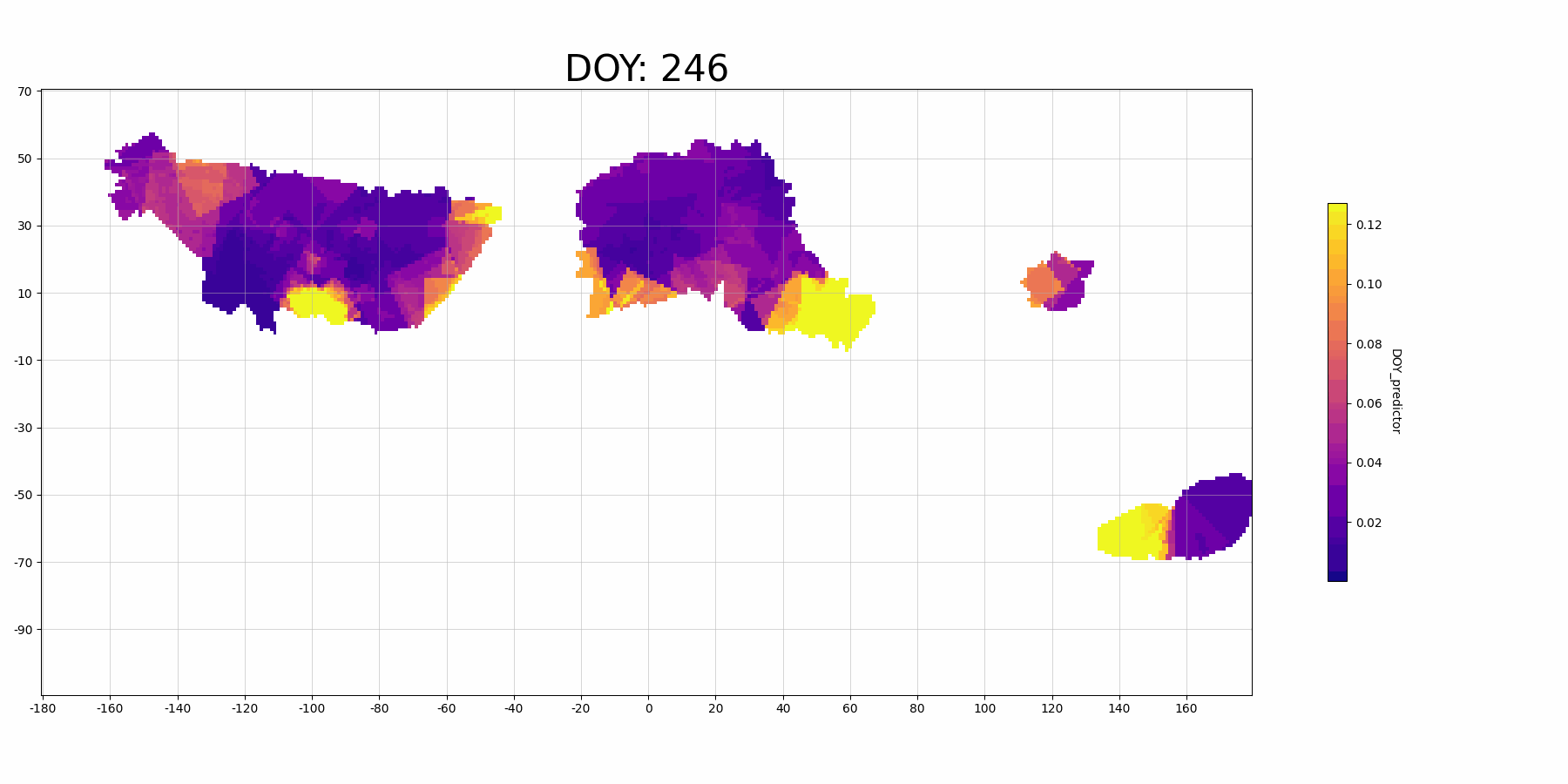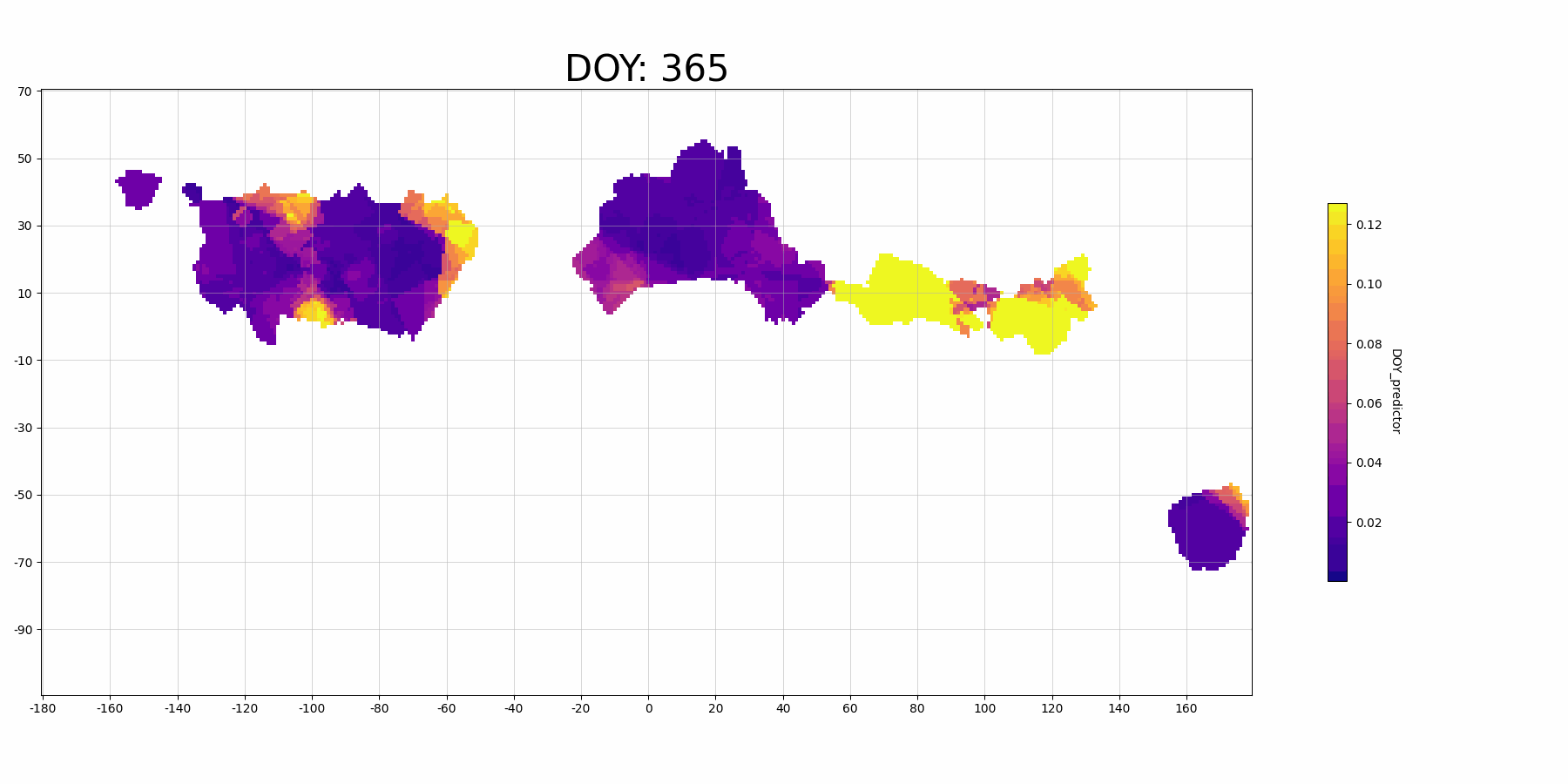
Focussing on North America (the large block on the left, longitude -160 to -40): The predictor DOY (day of year) seems to be a more important predictor during spring (30-150 DOY) than other time periods, which makes sense because spring migration is highly scheduled. And there is also higher importance in the middle migration flyway. This may indicate that there are more migrant Mallard population (since they follow the time heavily) in this regions.
Plot uncertainty in training¶
stemflow support calculating the variation of prediction across ensembles. It represent the uncertainty of making this prediction based on the surrounding information.
# calculate mean and standard deviation in abundance prediction
pred_mean, pred_std = model.predict(X_train, return_std=True, verbosity=1, n_jobs=1)
Predicting: 100%|██████████| 10/10 [01:11<00:00, 7.12s/it]
Next, we can plot these uncertainties. We visualize them by first aggregate them to hexagons using h3pandas package (you may install it if you haven't):
# make error_df
error_df = X_train[['longitude', 'latitude']]
error_df.columns = ['lng', 'lat']
error_df['pred_std'] = pred_std
error_df['log_pred_std'] = np.log(pred_std+1)
# Aggregate error to hexagon
import h3pandas # You can also use other aggregation method if you don't want to install h3pandas
H_level = 3
error_df = error_df.h3.geo_to_h3(H_level)
error_df = error_df.reset_index(drop=False).groupby(f'h3_0{H_level}').mean()
error_df = error_df.h3.h3_to_geo_boundary()
# plot mean uncertainty in hexagon
EU_error_df = error_df.query(
'-30<=lng<=50 & 20<=lat<=80'
)
EU_error_df.plot('log_pred_std', legend=True, legend_kwds={'shrink':0.7}, vmax = EU_error_df['log_pred_std'].quantile(0.9))
plt.grid(alpha=0.3)
plt.title('Log standard deviation in estimated mean abundance')
plt.xlabel('longitude')
plt.ylabel('latitude')
plt.show()
We see a higher relative prediction uncertainty in western Europe than eastern Europe.
Likewise, you can also aggregate the pred_std based on temporal features:
# make error_df
error_df = X_train[['longitude', 'latitude','DOY']]
error_df['log_pred_std'] = np.log(pred_std+1)
error_df.groupby('DOY')['log_pred_std'].mean().plot()
plt.ylabel('log_pred_std')
plt.show()
Interestingly, the prediction errors are the highest during spring and fall, when Mallards migrate, and lower during the summer and winter. It means the challenge of catching the abundance heterogeneity is larger during migration seasons.
Save model¶
If you are not using lazyloading (check model.lazy_loading), you can use pickle to save the model:
with open('./01.demo_adastem_model.pkl','wb') as f:
pickle.dump(model, f)
To load the model, do:
with open('./01.demo_adastem_model.pkl','rb') as f:
model = pickle.load(f)
This simple pickling only works for models with lazy_loading=False
If you are using lazy_loading=True, use:
model.save(tar_gz_file='./my_model.tar.gz', remove_temporary_file=True)
# After removing temporary files, you can no longer access to the ensembles saved on disk!
# Instead, they are in the .tar.gz file no2.
# Load model
model = AdaSTEM.load(tar_gz_file='./my_model.tar.gz', new_lazy_loading_path='./new_lazyloading_ensemble_folder', remove_original_file=False)
# If not specifying target_lazyloading_path, a random folder will be made under your current working directory.
model.lazy_loading_dir # notice that your lazy loading folder now is changed!
'new_lazyloading_ensemble_folder'
Make sure you use the same version of stemflow for write and load models
Evaluation¶
Now, we evaluate our overall model performance on the held-out test set:
pred = model.predict(X_test, verbosity=1)
Predicting: 100%|██████████| 10/10 [01:14<00:00, 7.41s/it]
The samples not predictable are output as np.nan:
perc = np.sum(np.isnan(pred.flatten()))/len(pred.flatten())
print(f'Percentage not predictable {round(perc*100, 2)}%')
Percentage not predictable 1.08%
AdaSTEM is relatively conservative compare to other models like Maxent, gradient boosting, or linear regression – It ameliorates the long-distance/long-range prediction problem. Spatiotemporal points are only predictable if they are covered by stixels defined during training. Therefore, distant points with no stixel coverage can not be predicted. Consequently, there are 0.56% percent of test samples that cannot be predicted by our model, based on the configuration that they have to have 7 ensembles covered (set when we created the model). These samples are too far way from the "knowledge zone" that we trained on in terms of space and time.
We evaluate the performance using various metrics implemented in AdaSTEM.eval_STEM_res method:
pred_df = pd.DataFrame({
'y_true':np.array(y_test).flatten(),
'y_pred':np.where(pred.flatten()<0, 0, pred.flatten())
}).dropna()
AdaSTEM.eval_STEM_res('hurdle', pred_df.y_true, pred_df.y_pred)
{'AUC': 0.8115519450595876,
'kappa': 0.34184247615390695,
'f1': 0.49449012525935604,
'precision': 0.3511218788196263,
'recall': 0.8357317263234454,
'average_precision': 0.320496619108254,
'Spearman_r': 0.46123808557541984,
'Pearson_r': 0.13693289373396436,
'R2': -0.025510676278269306,
'MAE': 4.833520909704486,
'MSE': 3663.3461640799533,
'poisson_deviance_explained': 0.22109384294027845}
The AUC reach 0.81, which is acceptable considering the quality of citizen science data. The recall is higher than precision, as expected. But R2 is low and MAE/MSE are high. This indicates that our model is good at classification, but the abundance output is not that good for downstream analysis. You may also argue that R2 is not appropriate here since it is a mixture of classification and regression question.
Compared to simple Hurdle model¶
What if we do not use AdaSTEM, instead, use a simple hurdle model with XGB base model?
model2 = Hurdle(classifier=XGBClassifier(tree_method='hist',random_state=42, verbosity = 0, n_jobs=1),
regressor=XGBRegressor(tree_method='hist',random_state=42, verbosity = 0, n_jobs=1))
model2.fit(X_train.drop(['longitude','latitude'], axis=1), y_train)
pred2 = model2.predict(X_test.drop(['longitude','latitude'], axis=1))
AdaSTEM.eval_STEM_res('hurdle', np.array(y_test).flatten(), np.where(pred2.flatten()<0, 0, pred2.flatten()))
{'AUC': 0.6090892200461546,
'kappa': 0.29060730357108033,
'f1': 0.3544066333996173,
'precision': 0.644514192139738,
'recall': 0.24439844760672705,
'average_precision': 0.28109960471554657,
'Spearman_r': 0.342922202734392,
'Pearson_r': 0.10530214595580313,
'R2': -0.0007916819661260011,
'MAE': 4.082427091361664,
'MSE': 3537.9218257720727,
'poisson_deviance_explained': 0.11952473790721874}
The untuned naive hurdle model did not out perform the AdaSTEM model, in terms of both classification metrics and regression metrics, which proves the advantages of using AdaSTEM framework.
Prediction¶
Now our model can show its full power: Making prediction
We first load the prediction set downloaded from https://figshare.com/articles/dataset/Predset_2020_csv/24124980 :
pred_set = pd.read_csv('./Predset_2020.csv')
For simplicity, we did not predict on the full set – Instead, we subsample the prediction set to 500 x 500 grids across the world, with one point per gird:
## reduce the prediction size
pred_set['lng_grid'] = np.digitize(
pred_set.longitude,
np.linspace(-180,180,500)
)
pred_set['lat_grid'] = np.digitize(
pred_set.latitude,
np.linspace(-90,90,500)
)
pred_set = pred_set.sample(frac=1, replace=False).groupby(['lng_grid','lat_grid']).first().reset_index(drop=True)
# pred_set = pred_set.drop(['lng_grid','lat_grid'], axis=1)
Then we can make our prediction. As mention before, the only dynamic feature in our model is DOY. To predict the abundance for each day, we only need to set the DOY to the target day. Additionally, we define the relative abundance prediction as
"A birder (number_observers=1)
who has observed 500 species (obsvr_species_count=500) in the past,
traveling 1km (Traveling=1; effort_distance_km=1)
within one hour (duration_minutes=60)
at 7:00 in the morning (time_observation_started_minute_of_day=420),
how many Mallard will they observe"
pred_df = []
for doy in tqdm(range(1,367)[::5]):
pred_set['DOY'] = doy
pred_set['duration_minutes'] = 60
pred_set['Traveling'] = 1
pred_set['Stationary'] = 0
pred_set['Area'] = 0
pred_set['effort_distance_km'] = 1
pred_set['number_observers'] = 1
pred_set['obsvr_species_count'] = 500
pred_set['time_observation_started_minute_of_day'] = 420
pred = model.predict(pred_set.fillna(-1), verbosity=0, n_jobs=5)
pred_df.append(pd.DataFrame({
'longitude':pred_set.longitude.values,
'latitude':pred_set.latitude.values,
'DOY':doy,
'pred':np.array(pred).flatten()
}))
not_p = np.sum(np.isnan(pred.flatten()))/len(pred.flatten())
# print(f'DOY {doy} Not predictable: {not_p*100}%')
0%| | 0/74 [00:00<?, ?it/s]
pred_df = pd.concat(pred_df, axis=0)
pred_df['pred'] = np.where(pred_df['pred']<0, 0, pred_df['pred'])
We set the prediction <0 to 0, because bird count cannot be negative.
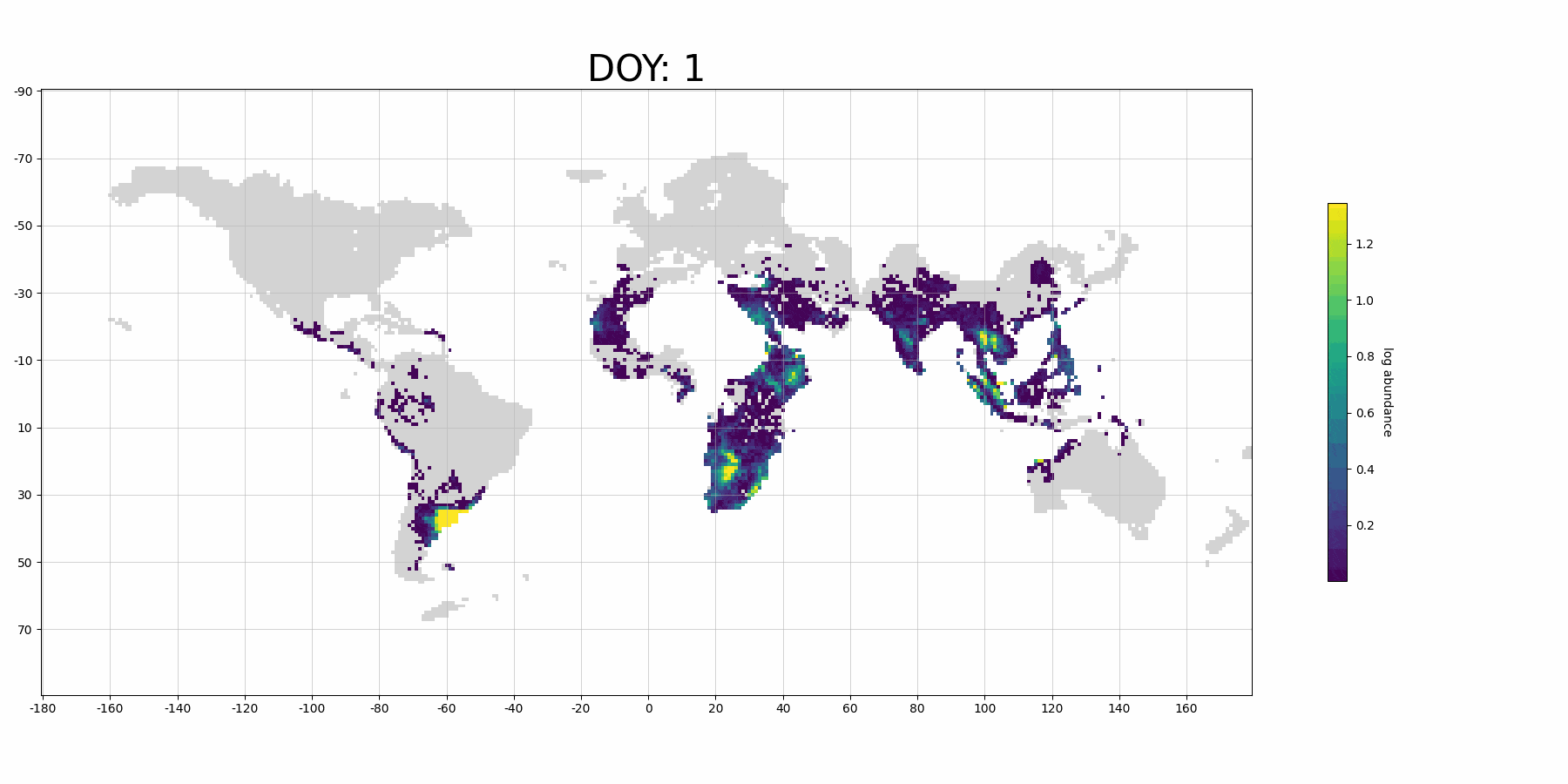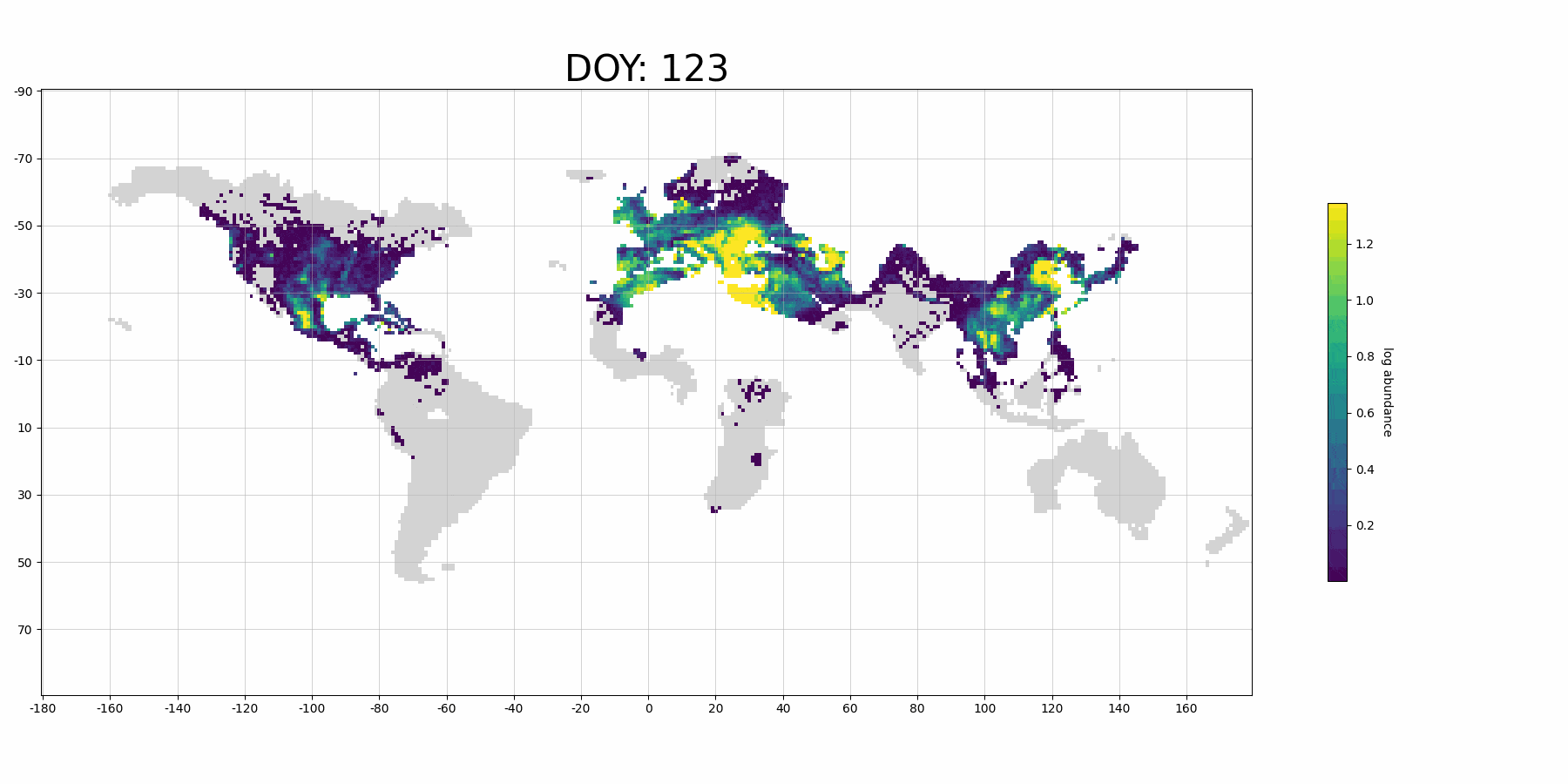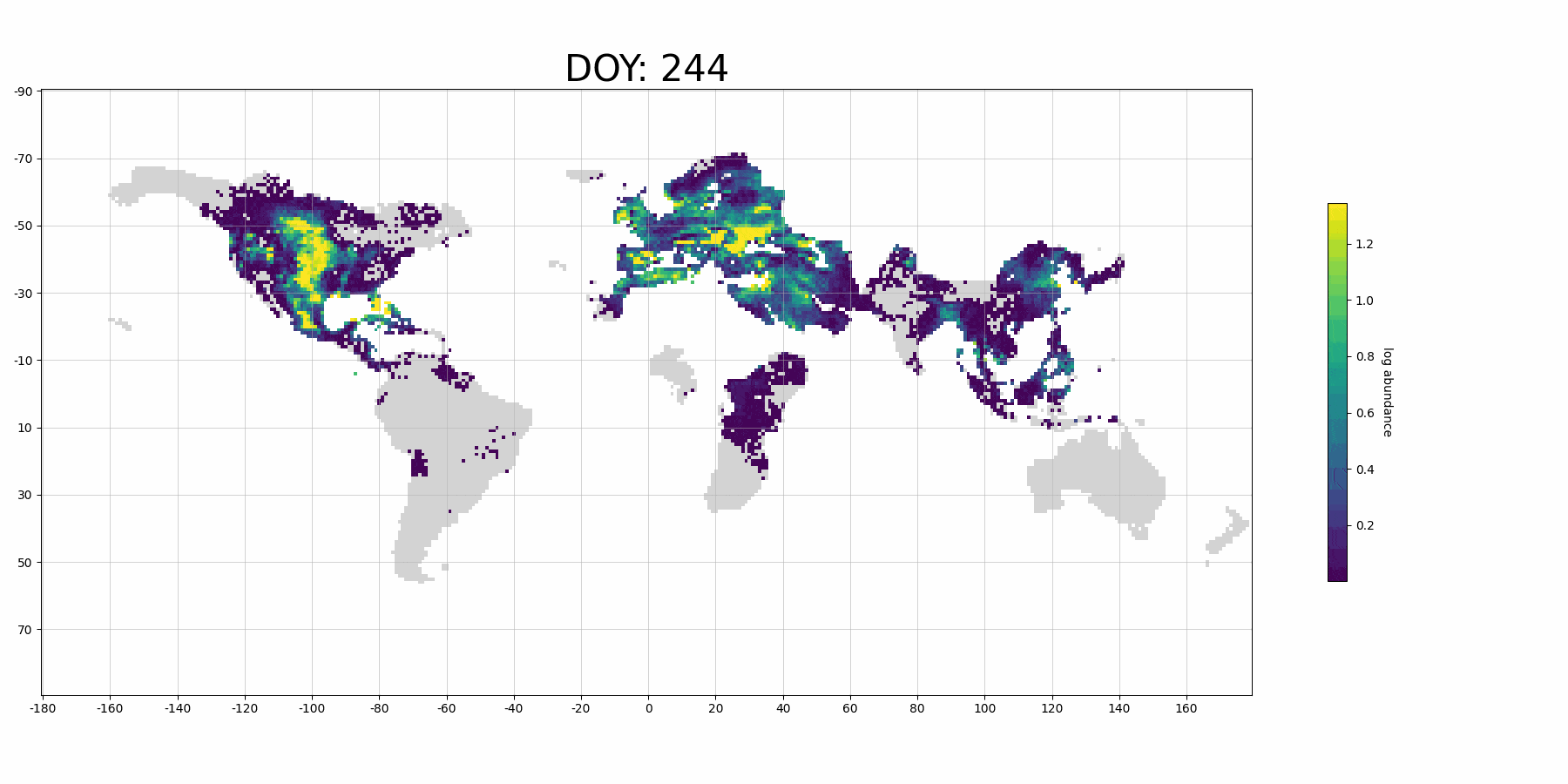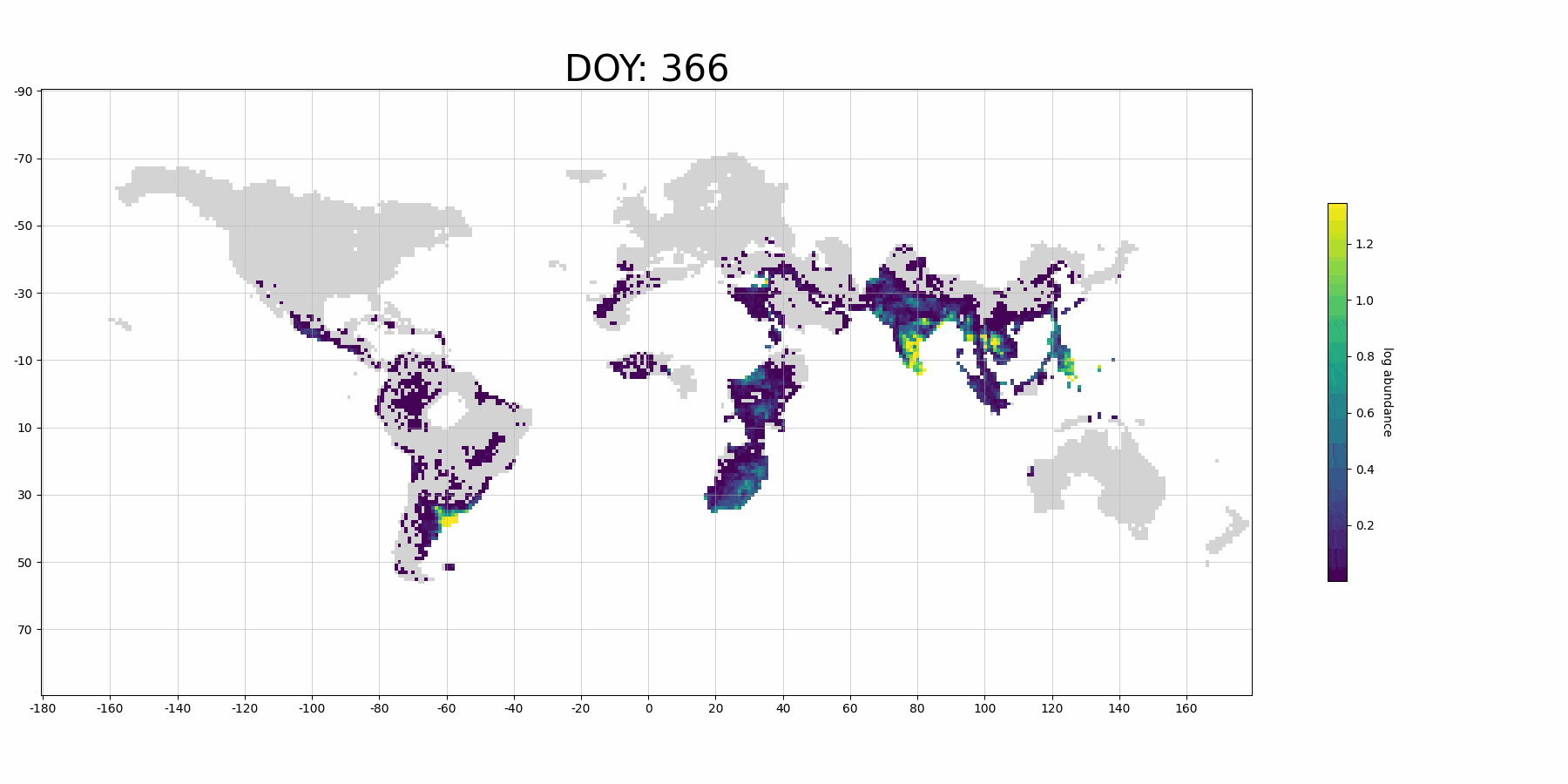
Next, we plot our prediction on map:
from stemflow.utils.plot_gif import make_sample_gif
make_sample_gif(pred_df, './pred_gif_demo.gif',
col='pred', log_scale = True,
Spatio1='longitude', Spatio2='latitude', Temporal1='DOY',
vmin=0.0001, vmax=pred_df['pred'].dropna().quantile(0.9),
cmap='viridis',
figsize=(18,9), xlims=(-180, 180), ylims=(-90,90), grid=True,
xtick_interval=20, ytick_interval=20,
lng_size = 360, lat_size = 180, dpi=100, fps=15)
Processing frame 74/74 Animation saved successfully!
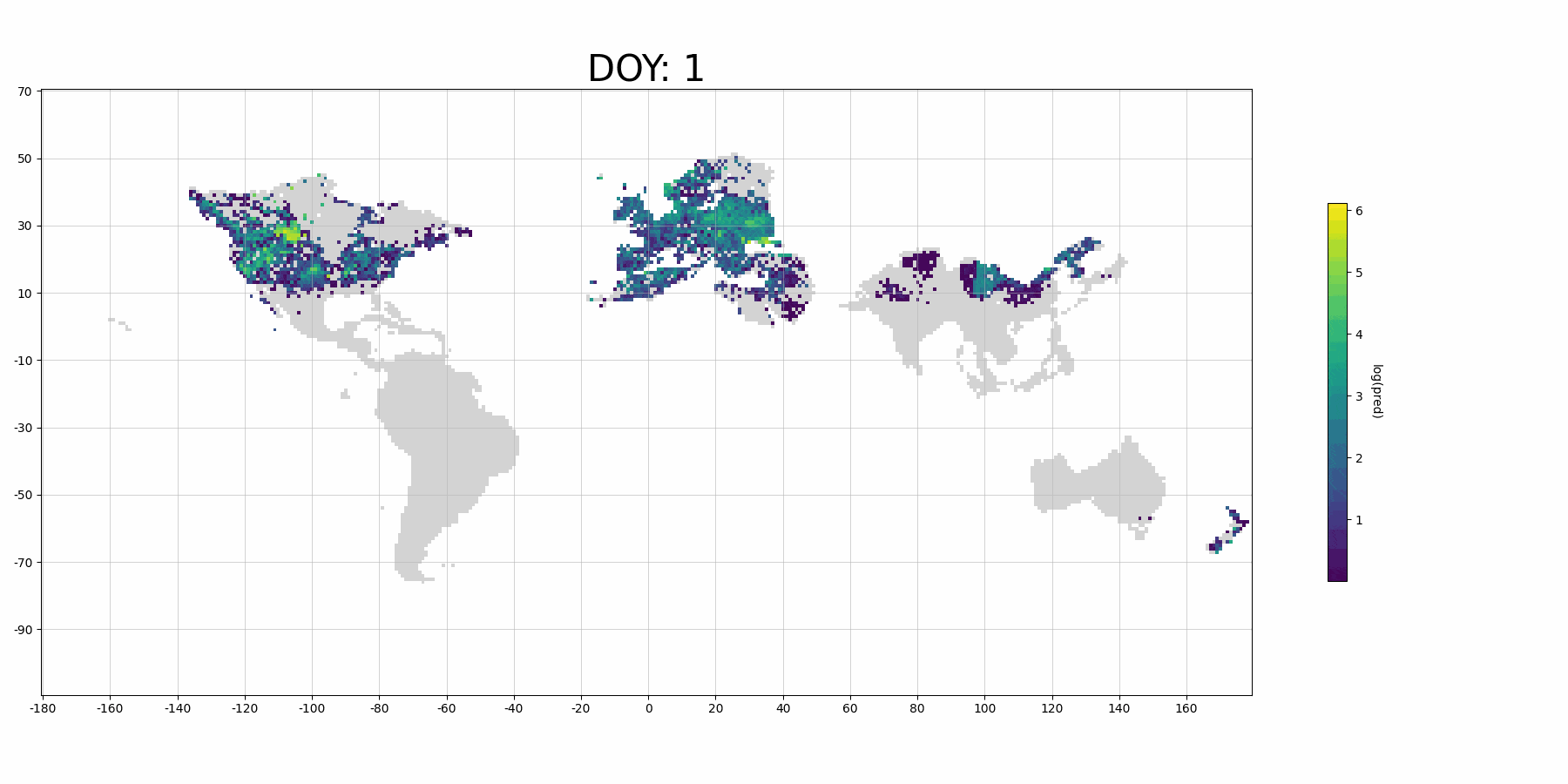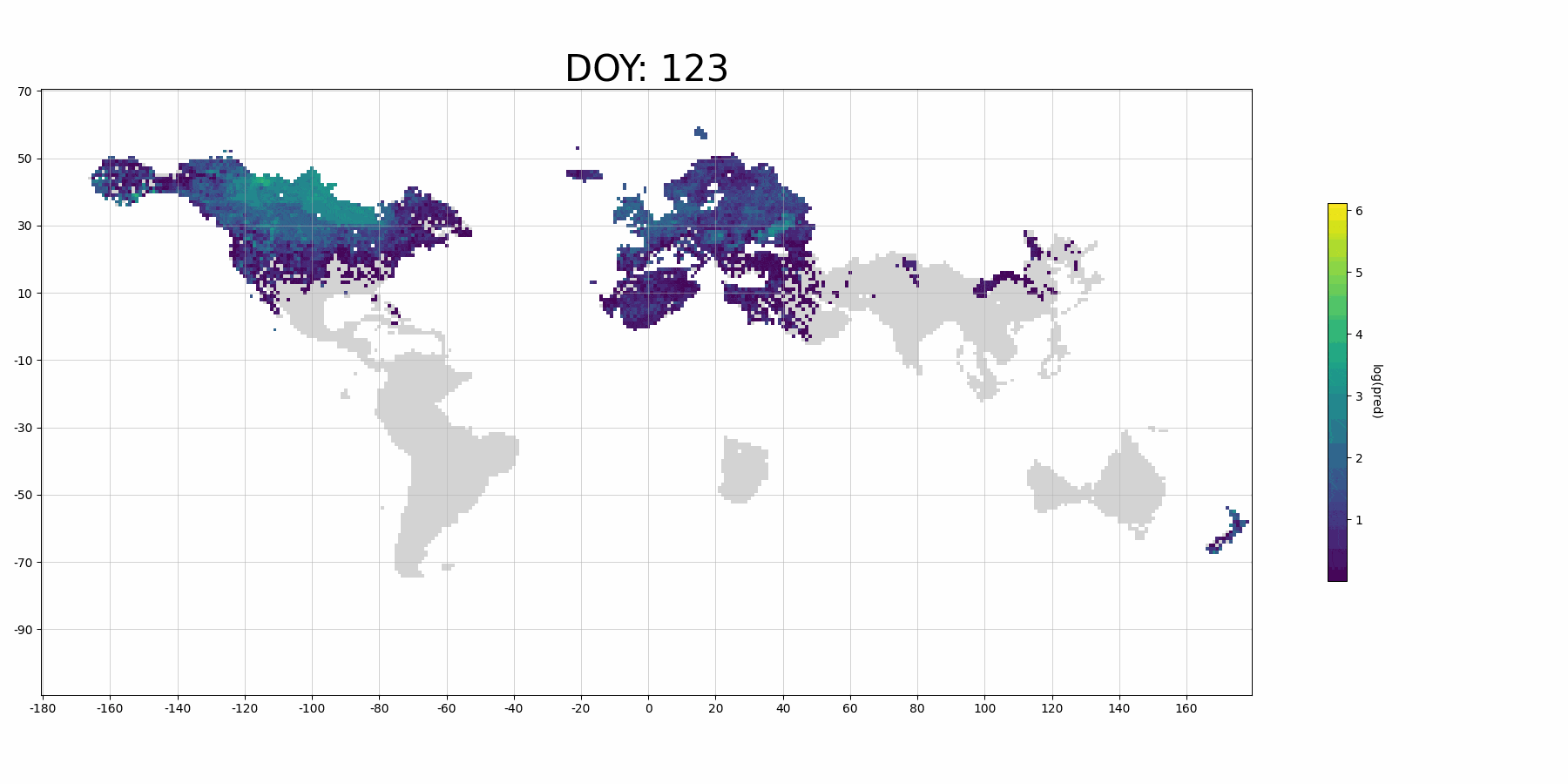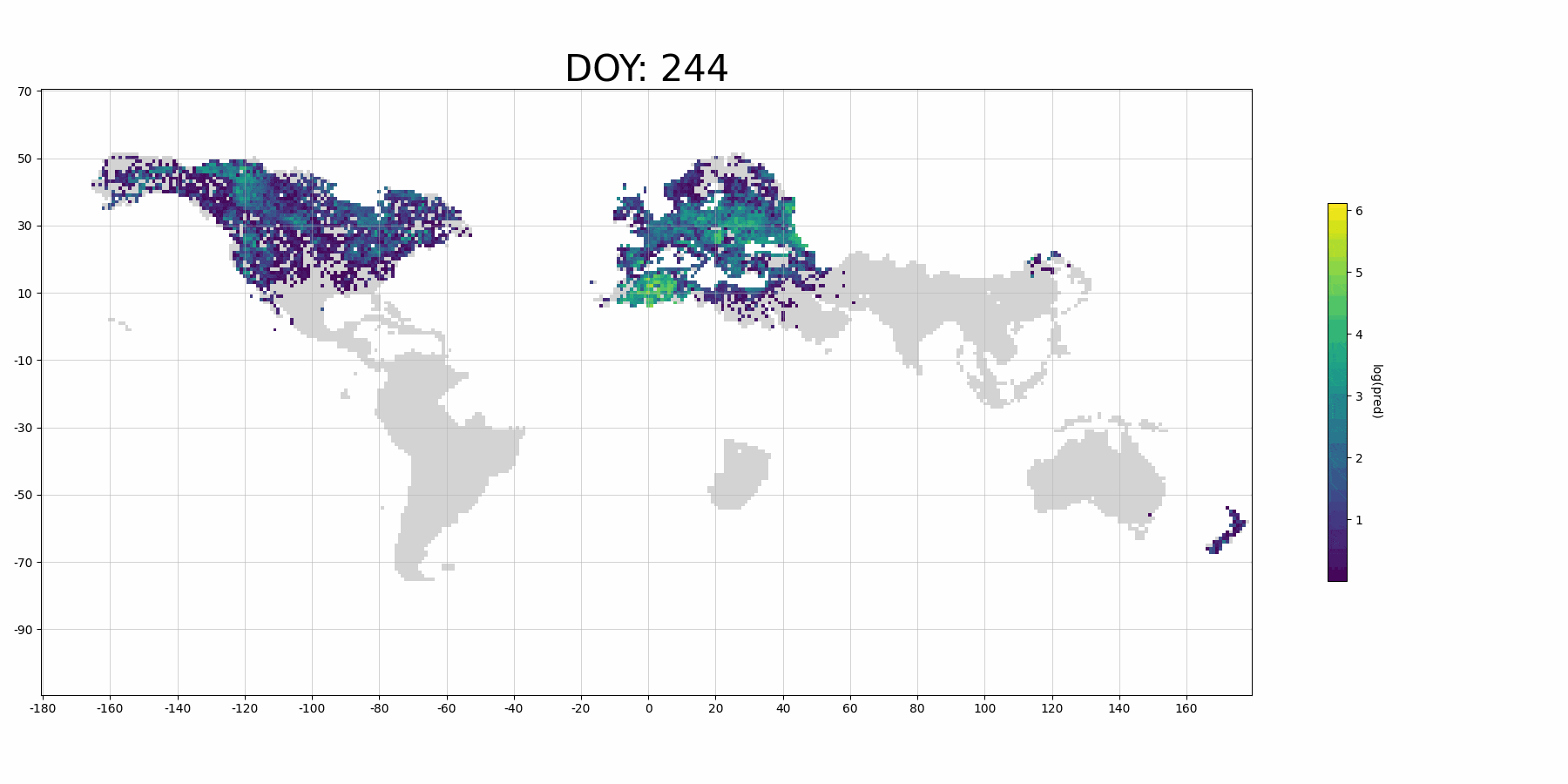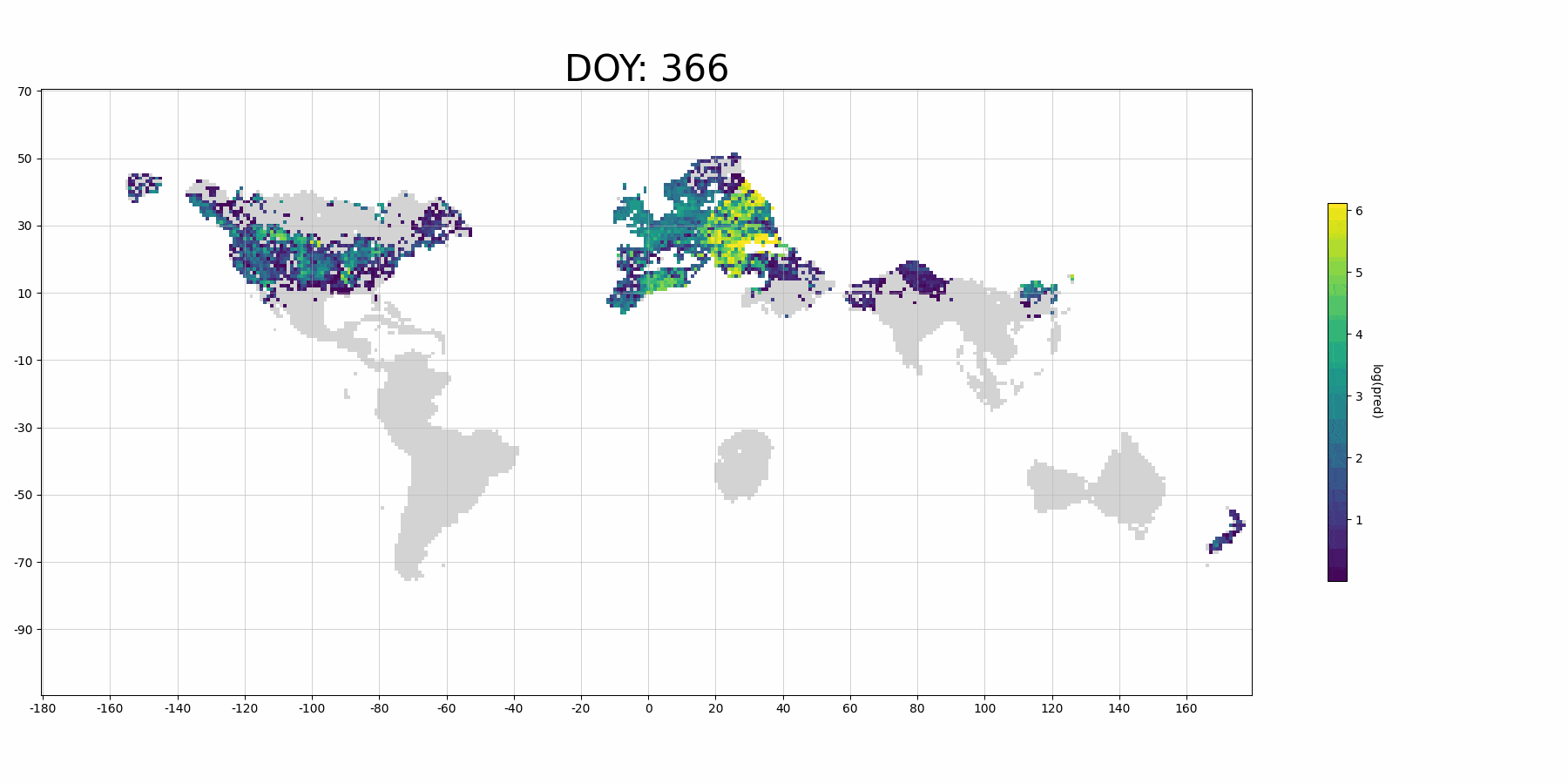
If you use more data for training and denser prediction set for visualization, it will probably look like (although it's for Barn Swallow):
Calculate memory usage¶
Finally, we are interested in how many disk space & memory our model consumed. stemflow is typically memory costly, especially when data volume and ensembles go up – the memory usage may also go up linearly. We do not currently provide solution for memory optimization. We well come PR to work on that.
# Calculate memory usage
model_size_G = round(os.path.getsize('./01.demo_adastem_model.pkl')/1024/1024/1024, 2)
training_data_memory_G = round(X.memory_usage().sum()/1024/1024/1024, 2)
# Calculate model info
from stemflow.lazyloading.lazyloading import LazyLoadingEstimator
true_hurdle_model_count = np.sum([1 if isinstance(model.model_dict[i], LazyLoadingEstimator) else 0 for i in model.model_dict])
dummy_model_count = len(model.model_dict) - true_hurdle_model_count
true_hurdle_model_perc = round(true_hurdle_model_count/len(model.model_dict) * 100, 2)
dummy_model_perc = round(dummy_model_count/len(model.model_dict) * 100, 2)
print(f"""
This AdaSTEM model have {len(model.model_dict)} trained based models in total.
Among them, {dummy_model_count} ({dummy_model_perc}%) are dummy models that always predict one class (because the input data labels are homogeneous).
Oppositely, {true_hurdle_model_count} ({true_hurdle_model_perc}%) are true hurdle models.
The input data consume {training_data_memory_G} G memory.
The model takes {model_size_G} G space on the disks.
""")
This AdaSTEM model have 9467 trained based models in total.
Among them, 2756 (29.11%) are dummy models that always predict one class (because the input data labels are homogeneous).
Oppositely, 6711 (70.89%) are true hurdle models.
The input data consume 0.15 G memory.
The model takes 0.01 G space on the disks.
Here we are using lazy_loading=True, which means the model will "dump itself" to the disk once it finishes training or after being used for prediction. This method keeps minimum memory but require some I/O reading and writing, so the prediction will be slower. If setting lazy_loading=False, the memory use will significantly increase.
Despite this economic aspect on the model side, the input training and prediction data will still be copied n_jobs times, since it will be used in each processor and different processors do not share memory. A more advanced option to avoid this and further reduce the memory load is using duckdb or parquet file path as input, so that each processor query those files independently in a lazy way, without loading everything into memory. For more information, see lazy loading and database query.
Other potentially useful functions¶
01. Obtaining spatial objects of stixels¶
Although for now stemflow doesn't have an internal function to obtain the spatial objects of stixels, one can realize it by:
from stemflow.utils.jitterrotation.jitterrotator import JitterRotator
from shapely.geometry import Polygon
import geopandas as gpd
# Remember to install shapely and geopandas if you haven't
# define a function
def geo_grid_geometry(line):
old_x, old_y = JitterRotator.inverse_jitter_rotate(
[line['stixel_calibration_point_transformed_left_bound'], line['stixel_calibration_point_transformed_left_bound'], line['stixel_calibration_point_transformed_right_bound'], line['stixel_calibration_point_transformed_right_bound']],
[line['stixel_calibration_point_transformed_lower_bound'], line['stixel_calibration_point_transformed_upper_bound'], line['stixel_calibration_point_transformed_upper_bound'], line['stixel_calibration_point_transformed_lower_bound']],
line['rotation'],
line['calibration_point_x_jitter'],
line['calibration_point_y_jitter'],
)
polygon = Polygon(list(zip(old_x, old_y)))
return polygon
# Make a geometry attribute for each stixel
model.ensemble_df['geometry'] = model.ensemble_df.apply(geo_grid_geometry, axis=1)
model.ensemble_df = gpd.GeoDataFrame(model.ensemble_df, geometry='geometry')
Which creates a spatial object for each stixel.
model.ensemble_df.plot(alpha=0.2)
<Axes: >
The stixels will stack on the temporal dimension. You may want to pick only one time range (since the spatial splitting pattern will be the same for all temporal windows in one ensemble).
model.ensemble_df[
(model.ensemble_df['DOY_start']>=90) & (model.ensemble_df['DOY_start']<120)
].plot(alpha=0.2)
<Axes: >
Which will look less messy.
You may also want to add scatter points on top of this:
fig,ax = plt.subplots()
model.ensemble_df[
(model.ensemble_df['DOY_start']>=90) & ((model.ensemble_df['DOY_start']<120))
].plot(alpha=0.2,ax=ax)
ax.scatter(
X_train['longitude'],
X_train['latitude'],
c='tab:orange',s=0.2,alpha=0.7
)
plt.show()
Concluding mark¶
Please open an issue if you have any question
Cheers!
from watermark import watermark
print(watermark())
print(watermark(packages="stemflow,numpy,scipy,pandas,xgboost,tqdm,matplotlib,h3pandas,geopandas,scikit-learn"))
Last updated: 2025-10-10T20:34:06.357488-05:00 Python implementation: CPython Python version : 3.11.11 IPython version : 8.31.0 Compiler : Clang 18.1.8 OS : Darwin Release : 24.6.0 Machine : arm64 Processor : arm CPU cores : 14 Architecture: 64bit stemflow : 1.1.5 numpy : 1.26.4 scipy : 1.16.1 pandas : 2.2.3 xgboost : 3.0.4 tqdm : 4.65.0 matplotlib : 3.10.0 h3pandas : 0.3.0 geopandas : 1.0.1 scikit-learn: 1.5.2